| Full name: pyrin domain containing 1 | Alias Symbol: ASC2|POP1 | ||
| Type: protein-coding gene | Cytoband: 16p11.2 | ||
| Entrez ID: 260434 | HGNC ID: HGNC:30261 | Ensembl Gene: ENSG00000169900 | OMIM ID: 615700 |
| Drug and gene relationship at DGIdb | |||
PYDC1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04621 | NOD-like receptor signaling pathway |
Expression of PYDC1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PYDC1 | 260434 | 243722_at | -0.0724 | 0.8196 | |
| GSE26886 | PYDC1 | 260434 | 243722_at | 0.2943 | 0.0868 | |
| GSE45670 | PYDC1 | 260434 | 243722_at | -0.0136 | 0.9109 | |
| GSE53622 | PYDC1 | 260434 | 10053 | 0.3531 | 0.0001 | |
| GSE53624 | PYDC1 | 260434 | 10053 | 0.5226 | 0.0000 | |
| GSE63941 | PYDC1 | 260434 | 243722_at | 0.2540 | 0.0844 | |
| GSE77861 | PYDC1 | 260434 | 243722_at | -0.1563 | 0.4910 | |
| GSE97050 | PYDC1 | 260434 | A_23_P407614 | 0.2541 | 0.2433 | |
| TCGA | PYDC1 | 260434 | RNAseq | 1.8630 | 0.1318 |
Upregulated datasets: 0; Downregulated datasets: 0.
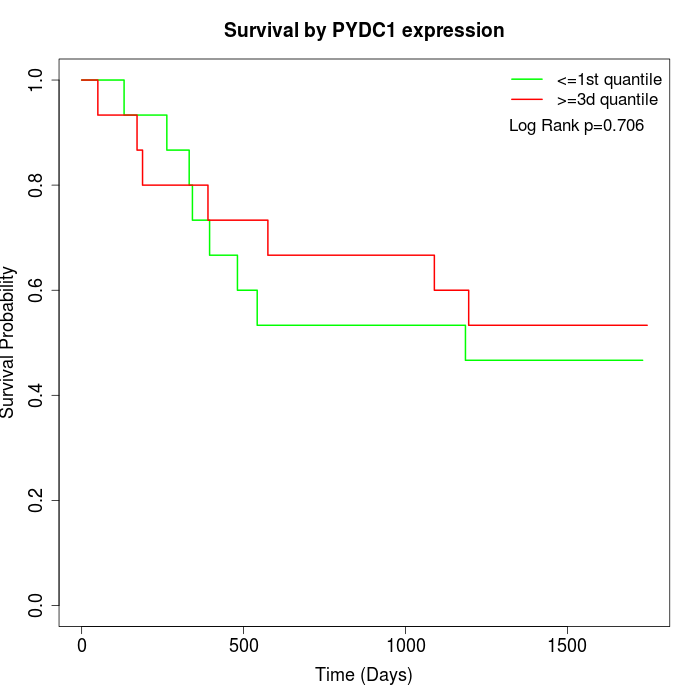
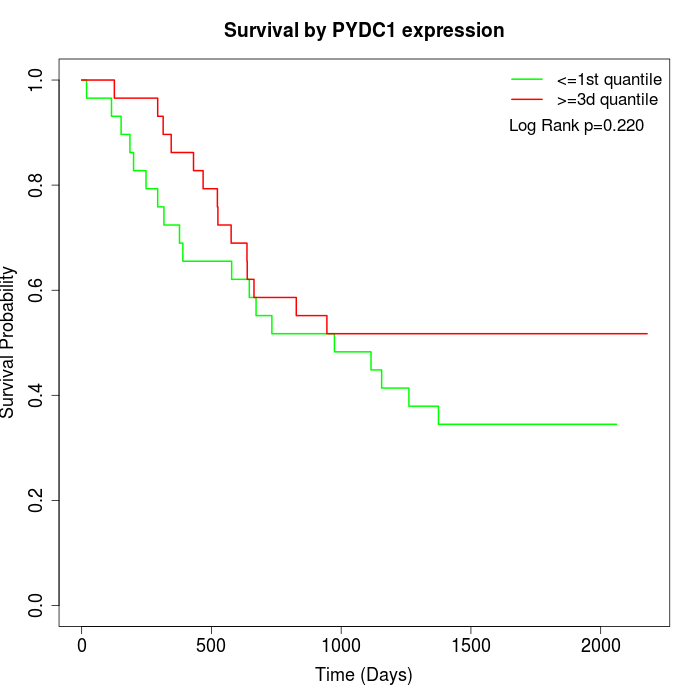
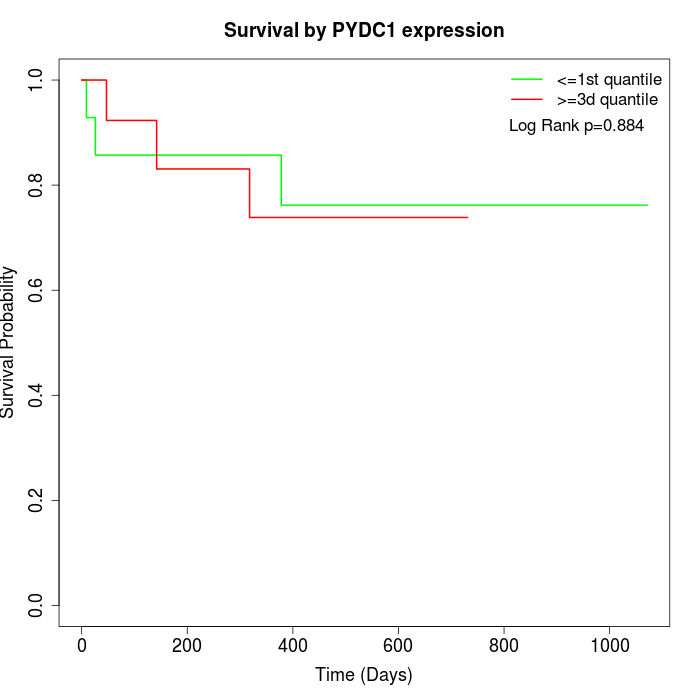
Survival by PYDC1 expression:
Note: Click image to view full size file.
Copy number change of PYDC1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PYDC1 | 260434 | 4 | 5 | 21 | |
| GSE20123 | PYDC1 | 260434 | 4 | 4 | 22 | |
| GSE43470 | PYDC1 | 260434 | 3 | 3 | 37 | |
| GSE46452 | PYDC1 | 260434 | 38 | 1 | 20 | |
| GSE47630 | PYDC1 | 260434 | 11 | 7 | 22 | |
| GSE54993 | PYDC1 | 260434 | 2 | 5 | 63 | |
| GSE54994 | PYDC1 | 260434 | 5 | 9 | 39 | |
| GSE60625 | PYDC1 | 260434 | 4 | 0 | 7 | |
| GSE74703 | PYDC1 | 260434 | 3 | 2 | 31 | |
| GSE74704 | PYDC1 | 260434 | 2 | 2 | 16 | |
| TCGA | PYDC1 | 260434 | 20 | 10 | 66 |
Total number of gains: 96; Total number of losses: 48; Total Number of normals: 344.
Somatic mutations of PYDC1:
Generating mutation plots.
Highly correlated genes for PYDC1:
Showing top 20/151 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PYDC1 | KRT74 | 0.74693 | 3 | 0 | 3 |
| PYDC1 | GNL1 | 0.711484 | 3 | 0 | 3 |
| PYDC1 | MAG | 0.696892 | 3 | 0 | 3 |
| PYDC1 | MPL | 0.69378 | 4 | 0 | 3 |
| PYDC1 | FOXB1 | 0.691168 | 3 | 0 | 3 |
| PYDC1 | NTN3 | 0.688892 | 3 | 0 | 3 |
| PYDC1 | GNRH2 | 0.688737 | 4 | 0 | 3 |
| PYDC1 | FLT4 | 0.682073 | 3 | 0 | 3 |
| PYDC1 | LTB4R2 | 0.680468 | 4 | 0 | 4 |
| PYDC1 | GRAP | 0.679431 | 3 | 0 | 3 |
| PYDC1 | MGAT5B | 0.669768 | 4 | 0 | 4 |
| PYDC1 | BEST3 | 0.66887 | 3 | 0 | 3 |
| PYDC1 | TMEM184A | 0.668174 | 4 | 0 | 3 |
| PYDC1 | ARMC5 | 0.666929 | 4 | 0 | 3 |
| PYDC1 | CHRNA4 | 0.663946 | 4 | 0 | 3 |
| PYDC1 | ZMYND10 | 0.662489 | 4 | 0 | 3 |
| PYDC1 | TBX10 | 0.66062 | 4 | 0 | 4 |
| PYDC1 | SNTG2 | 0.659686 | 3 | 0 | 3 |
| PYDC1 | PURB | 0.659136 | 3 | 0 | 3 |
| PYDC1 | TUT1 | 0.647747 | 3 | 0 | 3 |
For details and further investigation, click here