| Full name: C-C motif chemokine ligand 22 | Alias Symbol: MDC|STCP-1|ABCD-1|DC/B-CK|A-152E5.1|MGC34554 | ||
| Type: protein-coding gene | Cytoband: 16q21 | ||
| Entrez ID: 6367 | HGNC ID: HGNC:10621 | Ensembl Gene: ENSG00000102962 | OMIM ID: 602957 |
| Drug and gene relationship at DGIdb | |||
CCL22 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04062 | Chemokine signaling pathway |
Expression of CCL22:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CCL22 | 6367 | 207861_at | 0.3135 | 0.6285 | |
| GSE20347 | CCL22 | 6367 | 207861_at | -0.1572 | 0.2171 | |
| GSE23400 | CCL22 | 6367 | 207861_at | -0.0617 | 0.1794 | |
| GSE26886 | CCL22 | 6367 | 207861_at | -0.4592 | 0.0182 | |
| GSE29001 | CCL22 | 6367 | 207861_at | -0.2192 | 0.1738 | |
| GSE38129 | CCL22 | 6367 | 207861_at | 0.1010 | 0.5666 | |
| GSE45670 | CCL22 | 6367 | 207861_at | 0.3876 | 0.0291 | |
| GSE53622 | CCL22 | 6367 | 71068 | -0.0546 | 0.8529 | |
| GSE53624 | CCL22 | 6367 | 71068 | -0.5119 | 0.0302 | |
| GSE63941 | CCL22 | 6367 | 207861_at | 0.3476 | 0.3001 | |
| GSE77861 | CCL22 | 6367 | 207861_at | -0.0950 | 0.4369 | |
| GSE97050 | CCL22 | 6367 | A_24_P313418 | 0.6463 | 0.0867 | |
| SRP007169 | CCL22 | 6367 | RNAseq | 0.4569 | 0.5216 | |
| SRP064894 | CCL22 | 6367 | RNAseq | 0.3758 | 0.3501 | |
| SRP133303 | CCL22 | 6367 | RNAseq | 0.4393 | 0.1173 | |
| SRP159526 | CCL22 | 6367 | RNAseq | -0.1199 | 0.8599 | |
| SRP193095 | CCL22 | 6367 | RNAseq | 0.1897 | 0.2314 | |
| SRP219564 | CCL22 | 6367 | RNAseq | 0.4988 | 0.5126 | |
| TCGA | CCL22 | 6367 | RNAseq | 0.6810 | 0.0010 |
Upregulated datasets: 0; Downregulated datasets: 0.
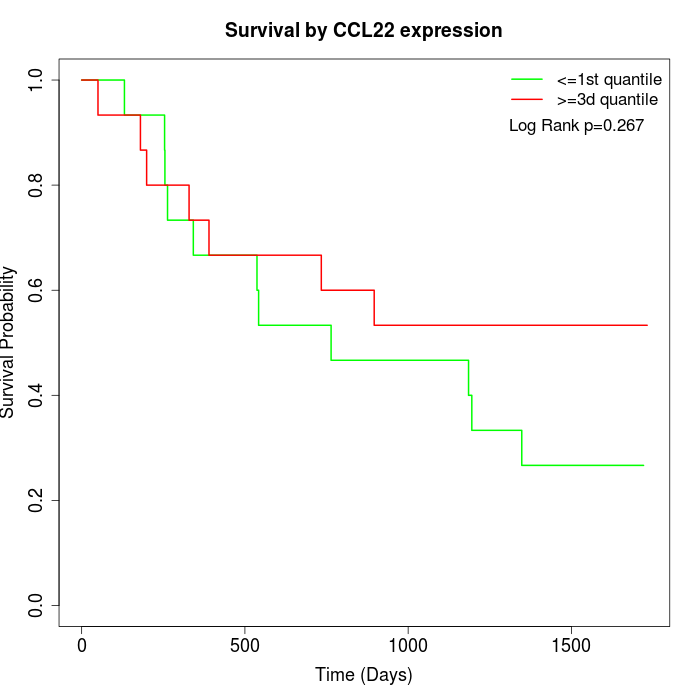
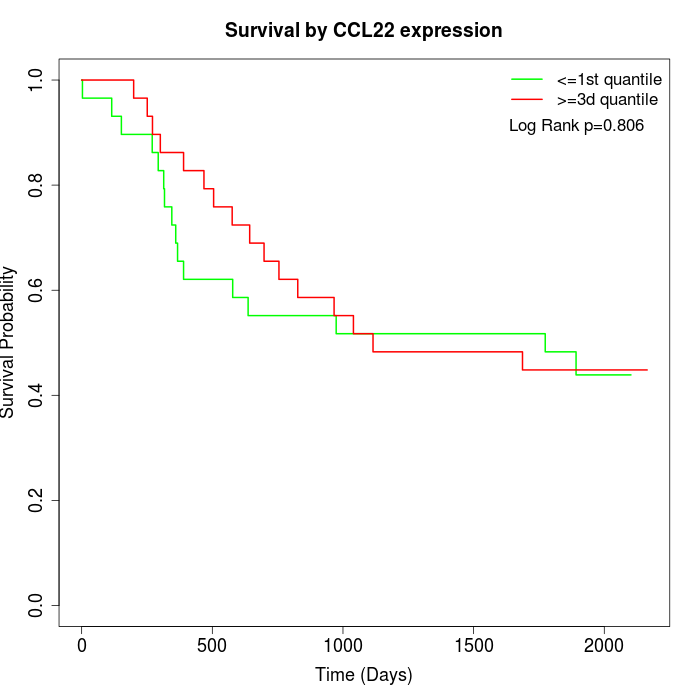
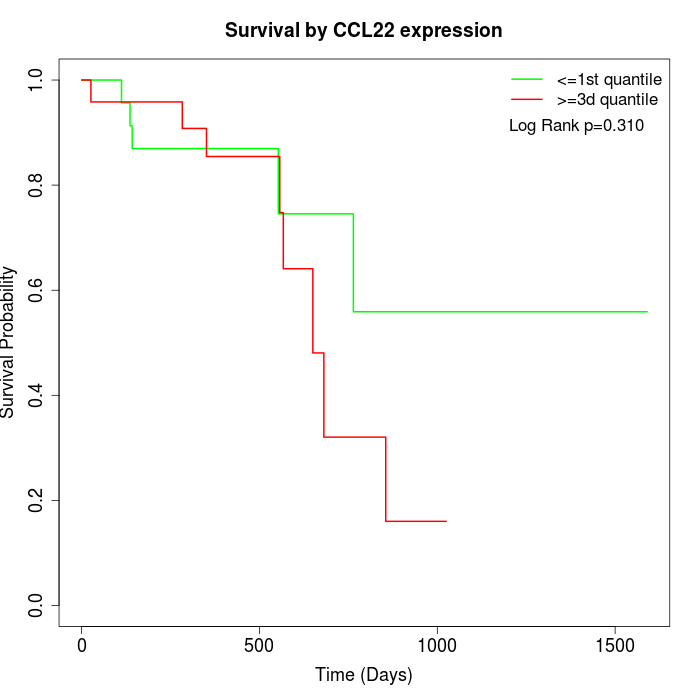
Survival by CCL22 expression:
Note: Click image to view full size file.
Copy number change of CCL22:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CCL22 | 6367 | 6 | 0 | 24 | |
| GSE20123 | CCL22 | 6367 | 6 | 1 | 23 | |
| GSE43470 | CCL22 | 6367 | 3 | 6 | 34 | |
| GSE46452 | CCL22 | 6367 | 38 | 1 | 20 | |
| GSE47630 | CCL22 | 6367 | 10 | 8 | 22 | |
| GSE54993 | CCL22 | 6367 | 2 | 4 | 64 | |
| GSE54994 | CCL22 | 6367 | 7 | 10 | 36 | |
| GSE60625 | CCL22 | 6367 | 4 | 0 | 7 | |
| GSE74703 | CCL22 | 6367 | 3 | 3 | 30 | |
| GSE74704 | CCL22 | 6367 | 5 | 0 | 15 | |
| TCGA | CCL22 | 6367 | 26 | 12 | 58 |
Total number of gains: 110; Total number of losses: 45; Total Number of normals: 333.
Somatic mutations of CCL22:
Generating mutation plots.
Highly correlated genes for CCL22:
Showing top 20/398 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CCL22 | DERL3 | 0.763898 | 3 | 0 | 3 |
| CCL22 | BIRC3 | 0.719962 | 3 | 0 | 3 |
| CCL22 | GALM | 0.712101 | 3 | 0 | 3 |
| CCL22 | PDE4B | 0.711467 | 3 | 0 | 3 |
| CCL22 | GPR37L1 | 0.711311 | 3 | 0 | 3 |
| CCL22 | PIM2 | 0.707224 | 5 | 0 | 4 |
| CCL22 | RITA1 | 0.703171 | 3 | 0 | 3 |
| CCL22 | TFAP4 | 0.692336 | 3 | 0 | 3 |
| CCL22 | MYO5C | 0.692209 | 3 | 0 | 3 |
| CCL22 | GIMAP4 | 0.684549 | 4 | 0 | 4 |
| CCL22 | CTRB2 | 0.683346 | 4 | 0 | 4 |
| CCL22 | ZNF358 | 0.67825 | 3 | 0 | 3 |
| CCL22 | S1PR2 | 0.676905 | 3 | 0 | 3 |
| CCL22 | DHDH | 0.676828 | 3 | 0 | 3 |
| CCL22 | PTPRU | 0.675659 | 4 | 0 | 3 |
| CCL22 | CLPP | 0.671187 | 3 | 0 | 3 |
| CCL22 | MAPRE3 | 0.668589 | 3 | 0 | 3 |
| CCL22 | INTS9 | 0.66673 | 3 | 0 | 3 |
| CCL22 | RBPJL | 0.663154 | 3 | 0 | 3 |
| CCL22 | E2F4 | 0.659771 | 4 | 0 | 4 |
For details and further investigation, click here