| Full name: C-C motif chemokine ligand 26 | Alias Symbol: MIP-4alpha|eotaxin-3|IMAC|MIP-4a|TSC-1 | ||
| Type: protein-coding gene | Cytoband: 7q11.23 | ||
| Entrez ID: 10344 | HGNC ID: HGNC:10625 | Ensembl Gene: ENSG00000006606 | OMIM ID: 604697 |
| Drug and gene relationship at DGIdb | |||
CCL26 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04062 | Chemokine signaling pathway |
Expression of CCL26:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CCL26 | 10344 | 223710_at | 0.5548 | 0.4798 | |
| GSE26886 | CCL26 | 10344 | 223710_at | 0.1688 | 0.4897 | |
| GSE45670 | CCL26 | 10344 | 223710_at | 0.9571 | 0.1500 | |
| GSE53622 | CCL26 | 10344 | 21205 | 0.9741 | 0.0000 | |
| GSE53624 | CCL26 | 10344 | 21205 | 1.1685 | 0.0000 | |
| GSE63941 | CCL26 | 10344 | 223710_at | -0.9846 | 0.4999 | |
| GSE77861 | CCL26 | 10344 | 223710_at | 0.3200 | 0.2594 | |
| GSE97050 | CCL26 | 10344 | A_23_P215484 | 0.1638 | 0.3993 | |
| SRP219564 | CCL26 | 10344 | RNAseq | 2.4083 | 0.0000 | |
| TCGA | CCL26 | 10344 | RNAseq | 2.9356 | 0.0000 |
Upregulated datasets: 3; Downregulated datasets: 0.
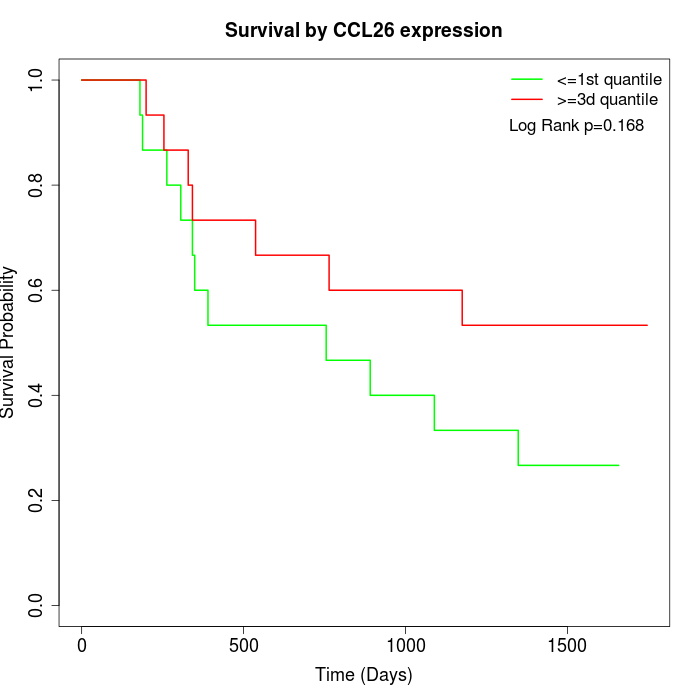
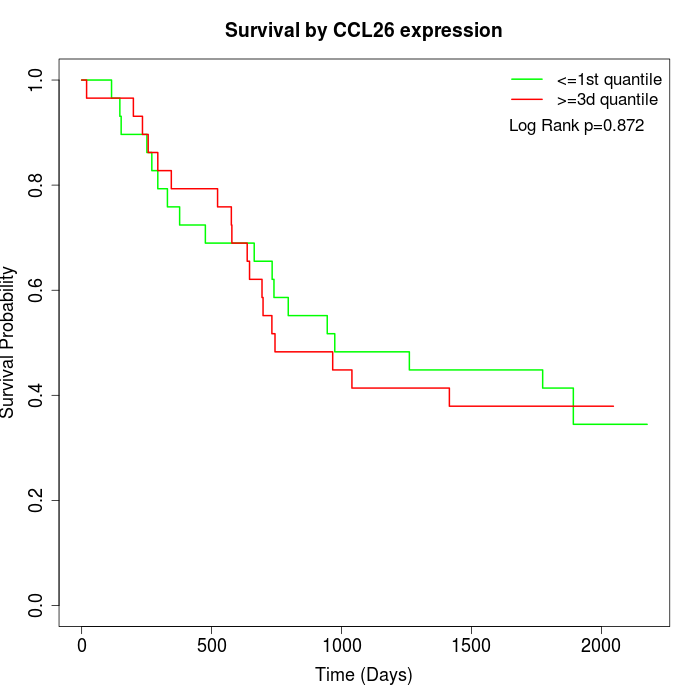
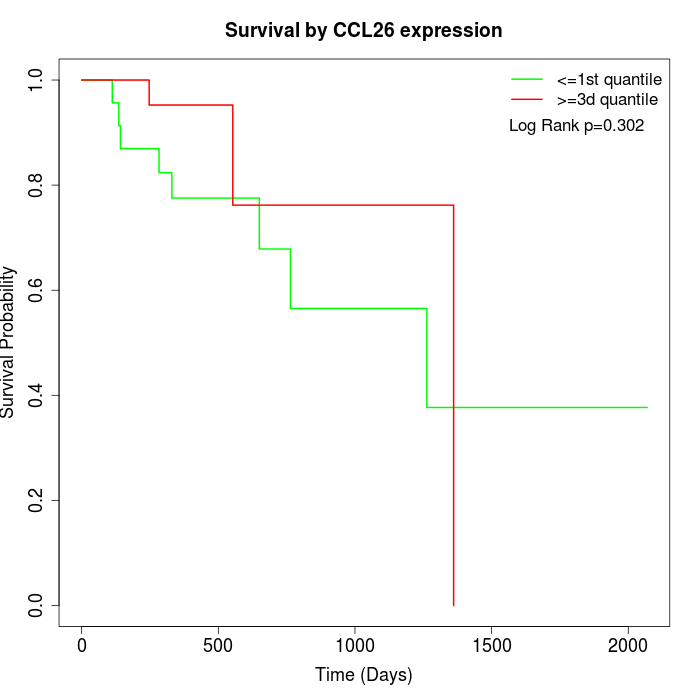
Survival by CCL26 expression:
Note: Click image to view full size file.
Copy number change of CCL26:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CCL26 | 10344 | 14 | 1 | 15 | |
| GSE20123 | CCL26 | 10344 | 14 | 1 | 15 | |
| GSE43470 | CCL26 | 10344 | 3 | 2 | 38 | |
| GSE46452 | CCL26 | 10344 | 12 | 0 | 47 | |
| GSE47630 | CCL26 | 10344 | 7 | 2 | 31 | |
| GSE54993 | CCL26 | 10344 | 2 | 7 | 61 | |
| GSE54994 | CCL26 | 10344 | 14 | 3 | 36 | |
| GSE60625 | CCL26 | 10344 | 0 | 0 | 11 | |
| GSE74703 | CCL26 | 10344 | 3 | 1 | 32 | |
| GSE74704 | CCL26 | 10344 | 8 | 1 | 11 | |
| TCGA | CCL26 | 10344 | 44 | 5 | 47 |
Total number of gains: 121; Total number of losses: 23; Total Number of normals: 344.
Somatic mutations of CCL26:
Generating mutation plots.
Highly correlated genes for CCL26:
Showing top 20/96 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CCL26 | ZNF236 | 0.829234 | 3 | 0 | 3 |
| CCL26 | KAZALD1 | 0.807425 | 3 | 0 | 3 |
| CCL26 | NOS3 | 0.771997 | 3 | 0 | 3 |
| CCL26 | NCR3 | 0.757626 | 3 | 0 | 3 |
| CCL26 | TLR9 | 0.74722 | 3 | 0 | 3 |
| CCL26 | SMOC1 | 0.722814 | 3 | 0 | 3 |
| CCL26 | ATG16L1 | 0.720793 | 3 | 0 | 3 |
| CCL26 | DPYSL4 | 0.720572 | 3 | 0 | 3 |
| CCL26 | ZFPM1 | 0.716729 | 3 | 0 | 3 |
| CCL26 | DUSP9 | 0.715701 | 3 | 0 | 3 |
| CCL26 | ORM1 | 0.708359 | 3 | 0 | 3 |
| CCL26 | IFT27 | 0.701307 | 4 | 0 | 4 |
| CCL26 | MMP19 | 0.696535 | 3 | 0 | 3 |
| CCL26 | CORT | 0.691982 | 3 | 0 | 3 |
| CCL26 | CECR2 | 0.690029 | 3 | 0 | 3 |
| CCL26 | KRTAP19-1 | 0.689377 | 4 | 0 | 3 |
| CCL26 | SPEF1 | 0.680203 | 3 | 0 | 3 |
| CCL26 | GCLM | 0.679988 | 5 | 0 | 4 |
| CCL26 | SLC39A5 | 0.67831 | 3 | 0 | 3 |
| CCL26 | PYCR1 | 0.676274 | 5 | 0 | 4 |
For details and further investigation, click here