| Full name: natural cytotoxicity triggering receptor 3 | Alias Symbol: 1C7|NKp30|CD337 | ||
| Type: protein-coding gene | Cytoband: 6p21.33 | ||
| Entrez ID: 259197 | HGNC ID: HGNC:19077 | Ensembl Gene: ENSG00000204475 | OMIM ID: 611550 |
| Drug and gene relationship at DGIdb | |||
NCR3 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04650 | Natural killer cell mediated cytotoxicity |
Expression of NCR3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NCR3 | 259197 | 210763_x_at | 0.1131 | 0.7915 | |
| GSE20347 | NCR3 | 259197 | 210763_x_at | 0.0525 | 0.3739 | |
| GSE23400 | NCR3 | 259197 | 211583_x_at | -0.1417 | 0.0003 | |
| GSE26886 | NCR3 | 259197 | 210763_x_at | 0.1635 | 0.2056 | |
| GSE29001 | NCR3 | 259197 | 211583_x_at | -0.1100 | 0.6106 | |
| GSE38129 | NCR3 | 259197 | 211010_s_at | -0.1510 | 0.0959 | |
| GSE45670 | NCR3 | 259197 | 211010_s_at | -0.0333 | 0.7502 | |
| GSE53622 | NCR3 | 259197 | 130320 | -0.0901 | 0.1827 | |
| GSE53624 | NCR3 | 259197 | 130320 | -0.1768 | 0.0051 | |
| GSE63941 | NCR3 | 259197 | 210763_x_at | 0.0668 | 0.6837 | |
| GSE77861 | NCR3 | 259197 | 210763_x_at | -0.0016 | 0.9908 | |
| GSE97050 | NCR3 | 259197 | A_33_P3344423 | 0.0455 | 0.8335 | |
| TCGA | NCR3 | 259197 | RNAseq | -0.2058 | 0.7631 |
Upregulated datasets: 0; Downregulated datasets: 0.
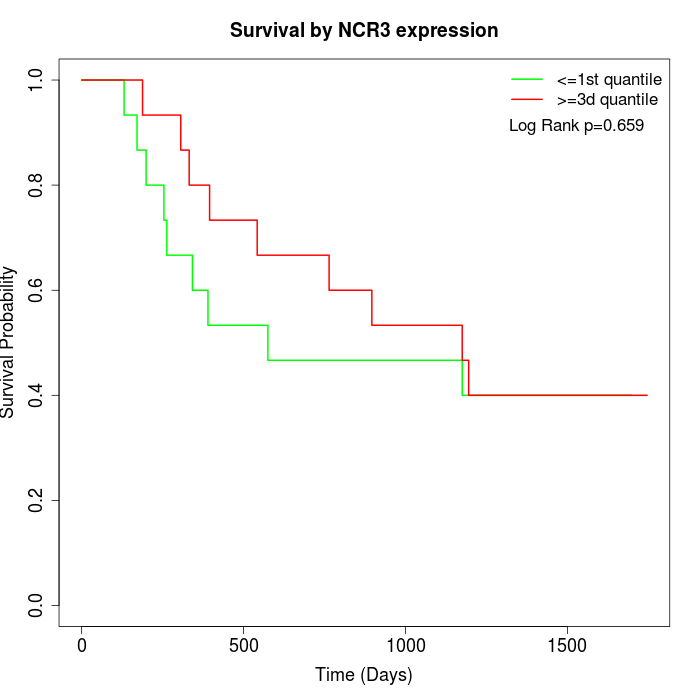
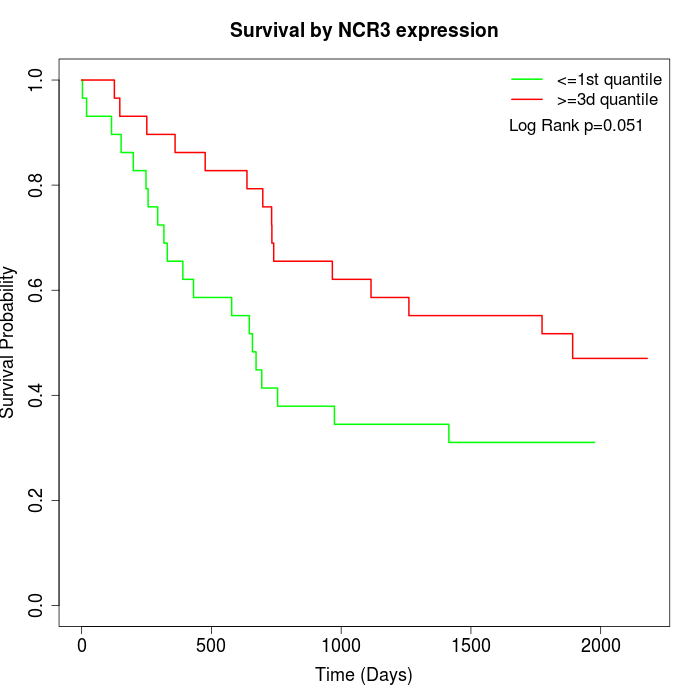
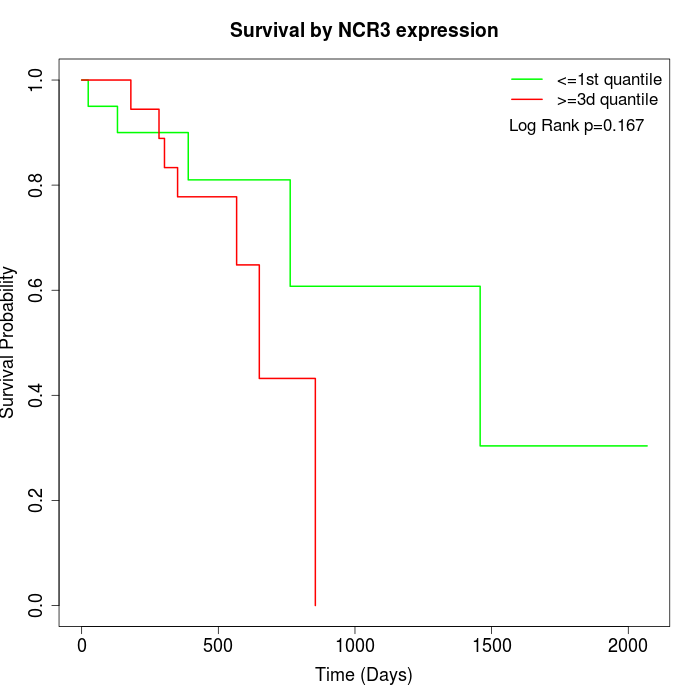
Survival by NCR3 expression:
Note: Click image to view full size file.
Copy number change of NCR3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NCR3 | 259197 | 5 | 1 | 24 | |
| GSE20123 | NCR3 | 259197 | 5 | 1 | 24 | |
| GSE43470 | NCR3 | 259197 | 6 | 3 | 34 | |
| GSE46452 | NCR3 | 259197 | 1 | 10 | 48 | |
| GSE47630 | NCR3 | 259197 | 7 | 5 | 28 | |
| GSE54993 | NCR3 | 259197 | 2 | 1 | 67 | |
| GSE54994 | NCR3 | 259197 | 11 | 4 | 38 | |
| GSE60625 | NCR3 | 259197 | 0 | 1 | 10 | |
| GSE74703 | NCR3 | 259197 | 6 | 1 | 29 | |
| GSE74704 | NCR3 | 259197 | 2 | 0 | 18 | |
| TCGA | NCR3 | 259197 | 16 | 16 | 64 |
Total number of gains: 61; Total number of losses: 43; Total Number of normals: 384.
Somatic mutations of NCR3:
Generating mutation plots.
Highly correlated genes for NCR3:
Showing top 20/963 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NCR3 | TRIM46 | 0.830938 | 3 | 0 | 3 |
| NCR3 | TLR9 | 0.777153 | 3 | 0 | 3 |
| NCR3 | HK3 | 0.770695 | 4 | 0 | 3 |
| NCR3 | ABCA4 | 0.763507 | 4 | 0 | 4 |
| NCR3 | THPO | 0.761773 | 3 | 0 | 3 |
| NCR3 | CELF4 | 0.75996 | 3 | 0 | 3 |
| NCR3 | CCL26 | 0.757626 | 3 | 0 | 3 |
| NCR3 | PRAMEF12 | 0.754341 | 4 | 0 | 3 |
| NCR3 | SLC18A3 | 0.749393 | 3 | 0 | 3 |
| NCR3 | FAM163A | 0.749234 | 3 | 0 | 3 |
| NCR3 | PALM3 | 0.748519 | 3 | 0 | 3 |
| NCR3 | FBXO15 | 0.743739 | 3 | 0 | 3 |
| NCR3 | PLA2G4D | 0.738886 | 3 | 0 | 3 |
| NCR3 | CCDC81 | 0.738176 | 3 | 0 | 3 |
| NCR3 | LHX9 | 0.733846 | 3 | 0 | 3 |
| NCR3 | FOXE3 | 0.726547 | 4 | 0 | 3 |
| NCR3 | DUSP9 | 0.722601 | 3 | 0 | 3 |
| NCR3 | ZIC1 | 0.719871 | 3 | 0 | 3 |
| NCR3 | EPHA10 | 0.719176 | 3 | 0 | 3 |
| NCR3 | CADPS | 0.716477 | 4 | 0 | 3 |
For details and further investigation, click here