| Full name: cytochrome P450 family 4 subfamily F member 22 | Alias Symbol: FLJ39501 | ||
| Type: protein-coding gene | Cytoband: 19p13.12 | ||
| Entrez ID: 126410 | HGNC ID: HGNC:26820 | Ensembl Gene: ENSG00000171954 | OMIM ID: 611495 |
| Drug and gene relationship at DGIdb | |||
Expression of CYP4F22:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CYP4F22 | 126410 | 244692_at | -1.1263 | 0.3522 | |
| GSE26886 | CYP4F22 | 126410 | 244692_at | -1.2039 | 0.0000 | |
| GSE45670 | CYP4F22 | 126410 | 244692_at | -0.2185 | 0.3915 | |
| GSE63941 | CYP4F22 | 126410 | 244692_at | 0.1765 | 0.2166 | |
| GSE77861 | CYP4F22 | 126410 | 244692_at | -0.6194 | 0.0476 | |
| SRP007169 | CYP4F22 | 126410 | RNAseq | -3.8026 | 0.0000 | |
| SRP008496 | CYP4F22 | 126410 | RNAseq | -2.3262 | 0.0150 | |
| SRP064894 | CYP4F22 | 126410 | RNAseq | -2.8270 | 0.0000 | |
| SRP133303 | CYP4F22 | 126410 | RNAseq | -3.7493 | 0.0000 | |
| TCGA | CYP4F22 | 126410 | RNAseq | -0.3667 | 0.4388 |
Upregulated datasets: 0; Downregulated datasets: 5.
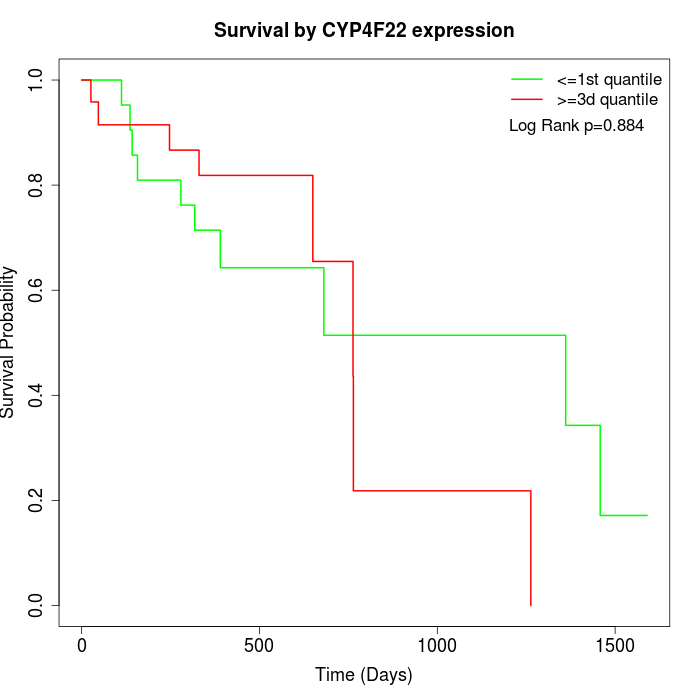
Survival by CYP4F22 expression:
Note: Click image to view full size file.
Copy number change of CYP4F22:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CYP4F22 | 126410 | 4 | 2 | 24 | |
| GSE20123 | CYP4F22 | 126410 | 3 | 1 | 26 | |
| GSE43470 | CYP4F22 | 126410 | 2 | 6 | 35 | |
| GSE46452 | CYP4F22 | 126410 | 47 | 1 | 11 | |
| GSE47630 | CYP4F22 | 126410 | 4 | 7 | 29 | |
| GSE54993 | CYP4F22 | 126410 | 15 | 4 | 51 | |
| GSE54994 | CYP4F22 | 126410 | 6 | 14 | 33 | |
| GSE60625 | CYP4F22 | 126410 | 9 | 0 | 2 | |
| GSE74703 | CYP4F22 | 126410 | 2 | 4 | 30 | |
| GSE74704 | CYP4F22 | 126410 | 0 | 1 | 19 | |
| TCGA | CYP4F22 | 126410 | 20 | 10 | 66 |
Total number of gains: 112; Total number of losses: 50; Total Number of normals: 326.
Somatic mutations of CYP4F22:
Generating mutation plots.
Highly correlated genes for CYP4F22:
Showing top 20/553 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CYP4F22 | CXCR2 | 0.80542 | 3 | 0 | 3 |
| CYP4F22 | RIOK3 | 0.777815 | 3 | 0 | 3 |
| CYP4F22 | EPB41L3 | 0.773259 | 3 | 0 | 3 |
| CYP4F22 | CAPN14 | 0.773134 | 3 | 0 | 3 |
| CYP4F22 | SLC16A6 | 0.769878 | 3 | 0 | 3 |
| CYP4F22 | CEACAM7 | 0.763211 | 3 | 0 | 3 |
| CYP4F22 | C15orf48 | 0.759851 | 3 | 0 | 3 |
| CYP4F22 | VPS4B | 0.757423 | 3 | 0 | 3 |
| CYP4F22 | CLCA4 | 0.754581 | 3 | 0 | 3 |
| CYP4F22 | PANK2 | 0.751131 | 3 | 0 | 3 |
| CYP4F22 | CLINT1 | 0.74891 | 3 | 0 | 3 |
| CYP4F22 | SPINK5 | 0.744658 | 3 | 0 | 3 |
| CYP4F22 | ENDOU | 0.740062 | 4 | 0 | 4 |
| CYP4F22 | CAPN5 | 0.739559 | 3 | 0 | 3 |
| CYP4F22 | PHACTR4 | 0.738249 | 3 | 0 | 3 |
| CYP4F22 | TIFA | 0.738198 | 3 | 0 | 3 |
| CYP4F22 | POF1B | 0.733434 | 3 | 0 | 3 |
| CYP4F22 | TST | 0.732292 | 3 | 0 | 3 |
| CYP4F22 | GDPD3 | 0.729411 | 4 | 0 | 4 |
| CYP4F22 | UPRT | 0.723867 | 3 | 0 | 3 |
For details and further investigation, click here