| Full name: gypsy retrotransposon integrase 1 | Alias Symbol: FLJ20125|GIN-1|TGIN1 | ||
| Type: protein-coding gene | Cytoband: 5q21.1 | ||
| Entrez ID: 54826 | HGNC ID: HGNC:25959 | Ensembl Gene: ENSG00000145723 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of GIN1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GIN1 | 54826 | 219467_at | -0.4558 | 0.2209 | |
| GSE20347 | GIN1 | 54826 | 219467_at | -0.4412 | 0.0030 | |
| GSE23400 | GIN1 | 54826 | 219467_at | -0.0376 | 0.4301 | |
| GSE26886 | GIN1 | 54826 | 219467_at | -0.5986 | 0.1083 | |
| GSE29001 | GIN1 | 54826 | 219467_at | -0.0915 | 0.7911 | |
| GSE38129 | GIN1 | 54826 | 219467_at | -0.2444 | 0.0505 | |
| GSE45670 | GIN1 | 54826 | 219467_at | -0.0338 | 0.8564 | |
| GSE53622 | GIN1 | 54826 | 36470 | -0.0959 | 0.2497 | |
| GSE53624 | GIN1 | 54826 | 43953 | 0.1116 | 0.2846 | |
| GSE63941 | GIN1 | 54826 | 219467_at | 0.4411 | 0.1535 | |
| GSE77861 | GIN1 | 54826 | 219467_at | -0.2396 | 0.2027 | |
| GSE97050 | GIN1 | 54826 | A_33_P3382910 | -0.1683 | 0.5842 | |
| SRP007169 | GIN1 | 54826 | RNAseq | -0.6322 | 0.0945 | |
| SRP064894 | GIN1 | 54826 | RNAseq | -0.1486 | 0.3403 | |
| SRP133303 | GIN1 | 54826 | RNAseq | 0.0490 | 0.7603 | |
| SRP159526 | GIN1 | 54826 | RNAseq | -0.4525 | 0.1253 | |
| SRP193095 | GIN1 | 54826 | RNAseq | -0.4985 | 0.0000 | |
| SRP219564 | GIN1 | 54826 | RNAseq | -0.7686 | 0.0006 | |
| TCGA | GIN1 | 54826 | RNAseq | -0.2979 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 0.
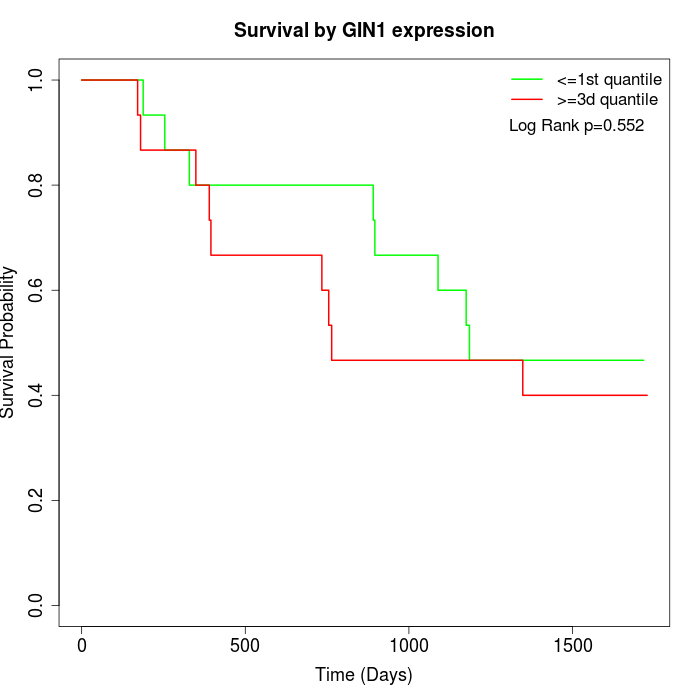
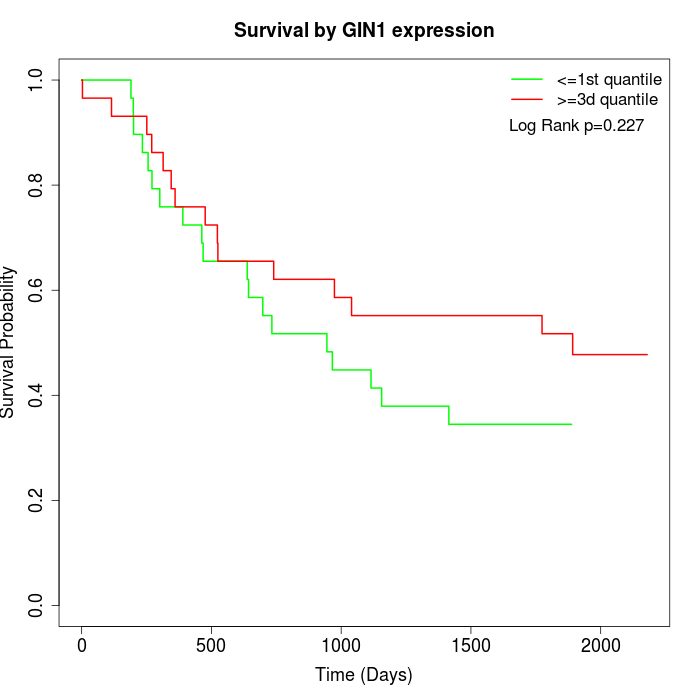
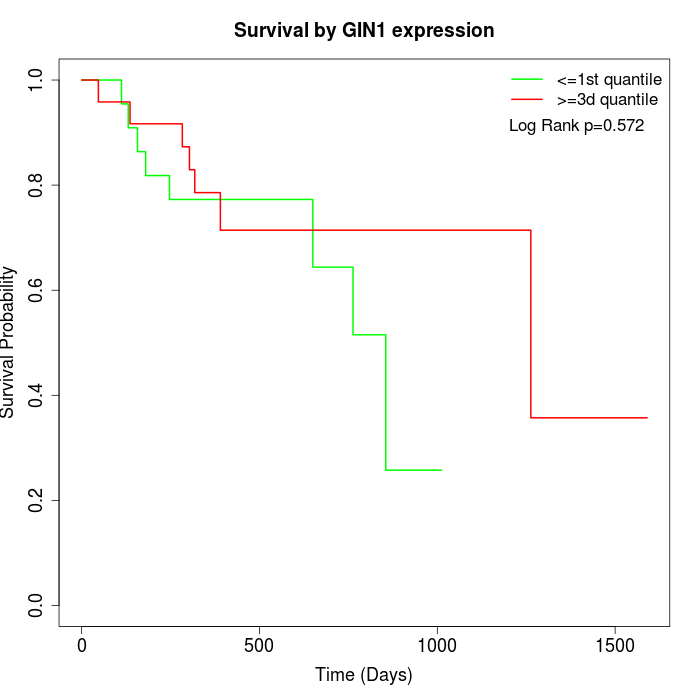
Survival by GIN1 expression:
Note: Click image to view full size file.
Copy number change of GIN1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GIN1 | 54826 | 1 | 13 | 16 | |
| GSE20123 | GIN1 | 54826 | 1 | 13 | 16 | |
| GSE43470 | GIN1 | 54826 | 3 | 9 | 31 | |
| GSE46452 | GIN1 | 54826 | 0 | 28 | 31 | |
| GSE47630 | GIN1 | 54826 | 0 | 20 | 20 | |
| GSE54993 | GIN1 | 54826 | 8 | 1 | 61 | |
| GSE54994 | GIN1 | 54826 | 1 | 17 | 35 | |
| GSE60625 | GIN1 | 54826 | 0 | 0 | 11 | |
| GSE74703 | GIN1 | 54826 | 2 | 7 | 27 | |
| GSE74704 | GIN1 | 54826 | 1 | 7 | 12 | |
| TCGA | GIN1 | 54826 | 2 | 43 | 51 |
Total number of gains: 19; Total number of losses: 158; Total Number of normals: 311.
Somatic mutations of GIN1:
Generating mutation plots.
Highly correlated genes for GIN1:
Showing top 20/389 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GIN1 | HOOK3 | 0.802085 | 3 | 0 | 3 |
| GIN1 | HDHD2 | 0.773231 | 3 | 0 | 3 |
| GIN1 | TAF7 | 0.770241 | 4 | 0 | 4 |
| GIN1 | ZCCHC8 | 0.770064 | 3 | 0 | 3 |
| GIN1 | BAG4 | 0.764042 | 3 | 0 | 3 |
| GIN1 | COPS5 | 0.763344 | 3 | 0 | 3 |
| GIN1 | FBXL3 | 0.75568 | 4 | 0 | 4 |
| GIN1 | DLD | 0.754985 | 4 | 0 | 4 |
| GIN1 | GPBP1 | 0.752261 | 5 | 0 | 5 |
| GIN1 | ECD | 0.747758 | 3 | 0 | 3 |
| GIN1 | ACSL1 | 0.746151 | 3 | 0 | 3 |
| GIN1 | AK3 | 0.742663 | 4 | 0 | 4 |
| GIN1 | WWP1 | 0.74033 | 3 | 0 | 3 |
| GIN1 | MFN1 | 0.734785 | 3 | 0 | 3 |
| GIN1 | CTDSPL | 0.72138 | 3 | 0 | 3 |
| GIN1 | MRPS36 | 0.71835 | 5 | 0 | 5 |
| GIN1 | DDX50 | 0.717257 | 3 | 0 | 3 |
| GIN1 | CLDND1 | 0.717135 | 3 | 0 | 3 |
| GIN1 | ADAM9 | 0.715281 | 3 | 0 | 3 |
| GIN1 | RPS6KB1 | 0.71262 | 3 | 0 | 3 |
For details and further investigation, click here