| Full name: CUGBP Elav-like family member 4 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 18q12.2 | ||
| Entrez ID: 56853 | HGNC ID: HGNC:14015 | Ensembl Gene: ENSG00000101489 | OMIM ID: 612679 |
| Drug and gene relationship at DGIdb | |||
Expression of CELF4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CELF4 | 56853 | 223653_x_at | 0.1136 | 0.7078 | |
| GSE26886 | CELF4 | 56853 | 223653_x_at | 0.0064 | 0.9667 | |
| GSE45670 | CELF4 | 56853 | 232719_at | 0.0630 | 0.5961 | |
| GSE53622 | CELF4 | 56853 | 19557 | 0.0222 | 0.7499 | |
| GSE53624 | CELF4 | 56853 | 19557 | 0.2938 | 0.0022 | |
| GSE63941 | CELF4 | 56853 | 232719_at | 0.0813 | 0.5951 | |
| GSE77861 | CELF4 | 56853 | 232719_at | -0.1690 | 0.1654 | |
| GSE97050 | CELF4 | 56853 | A_33_P3535972 | 0.4097 | 0.2564 | |
| SRP133303 | CELF4 | 56853 | RNAseq | -0.3718 | 0.0905 | |
| SRP219564 | CELF4 | 56853 | RNAseq | 1.0267 | 0.1116 | |
| TCGA | CELF4 | 56853 | RNAseq | -1.7350 | 0.0003 |
Upregulated datasets: 0; Downregulated datasets: 1.
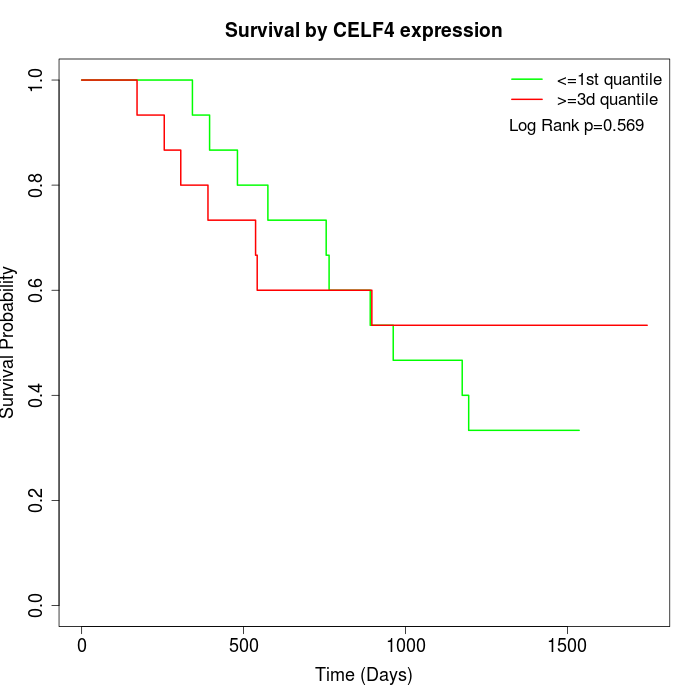
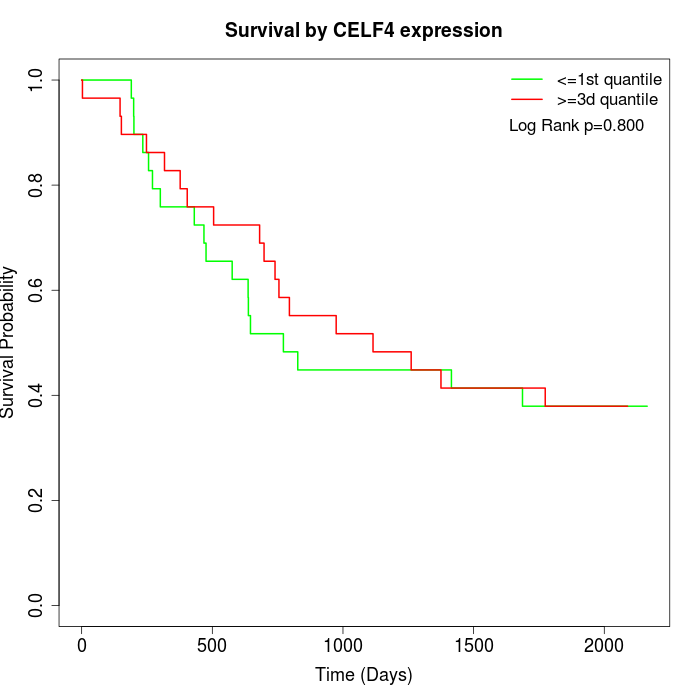
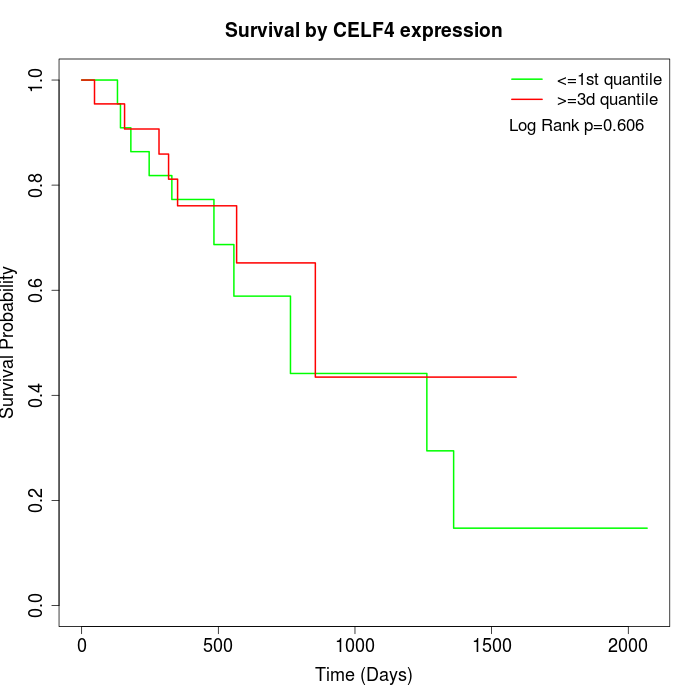
Survival by CELF4 expression:
Note: Click image to view full size file.
Copy number change of CELF4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CELF4 | 56853 | 1 | 6 | 23 | |
| GSE20123 | CELF4 | 56853 | 1 | 6 | 23 | |
| GSE43470 | CELF4 | 56853 | 0 | 3 | 40 | |
| GSE46452 | CELF4 | 56853 | 1 | 25 | 33 | |
| GSE47630 | CELF4 | 56853 | 5 | 20 | 15 | |
| GSE54993 | CELF4 | 56853 | 8 | 1 | 61 | |
| GSE54994 | CELF4 | 56853 | 1 | 16 | 36 | |
| GSE60625 | CELF4 | 56853 | 0 | 4 | 7 | |
| GSE74703 | CELF4 | 56853 | 0 | 3 | 33 | |
| GSE74704 | CELF4 | 56853 | 0 | 5 | 15 | |
| TCGA | CELF4 | 56853 | 14 | 40 | 42 |
Total number of gains: 31; Total number of losses: 129; Total Number of normals: 328.
Somatic mutations of CELF4:
Generating mutation plots.
Highly correlated genes for CELF4:
Showing top 20/223 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CELF4 | KRT25 | 0.831091 | 3 | 0 | 3 |
| CELF4 | MYL4 | 0.7887 | 3 | 0 | 3 |
| CELF4 | ISLR2 | 0.774873 | 3 | 0 | 3 |
| CELF4 | CHD5 | 0.773269 | 3 | 0 | 3 |
| CELF4 | ZFHX2 | 0.761357 | 3 | 0 | 3 |
| CELF4 | NCR3 | 0.75996 | 3 | 0 | 3 |
| CELF4 | FAM71E2 | 0.75418 | 3 | 0 | 3 |
| CELF4 | RGL4 | 0.753464 | 3 | 0 | 3 |
| CELF4 | SHC3 | 0.750074 | 3 | 0 | 3 |
| CELF4 | BEST3 | 0.747876 | 3 | 0 | 3 |
| CELF4 | DNAH6 | 0.747694 | 3 | 0 | 3 |
| CELF4 | NLRP5 | 0.74719 | 3 | 0 | 3 |
| CELF4 | IGFN1 | 0.743417 | 3 | 0 | 3 |
| CELF4 | CDHR5 | 0.737826 | 3 | 0 | 3 |
| CELF4 | ADAMTS7 | 0.737589 | 4 | 0 | 3 |
| CELF4 | OR1F1 | 0.736631 | 3 | 0 | 3 |
| CELF4 | ESPNL | 0.735343 | 4 | 0 | 3 |
| CELF4 | GPR17 | 0.733454 | 3 | 0 | 3 |
| CELF4 | CD70 | 0.733389 | 3 | 0 | 3 |
| CELF4 | STK32C | 0.733083 | 3 | 0 | 3 |
For details and further investigation, click here