| Full name: chromogranin A | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 14q32.12 | ||
| Entrez ID: 1113 | HGNC ID: HGNC:1929 | Ensembl Gene: ENSG00000100604 | OMIM ID: 118910 |
| Drug and gene relationship at DGIdb | |||
Expression of CHGA:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CHGA | 1113 | 204697_s_at | -0.1460 | 0.7208 | |
| GSE20347 | CHGA | 1113 | 204697_s_at | -0.1809 | 0.0111 | |
| GSE23400 | CHGA | 1113 | 204697_s_at | -0.1706 | 0.0001 | |
| GSE26886 | CHGA | 1113 | 204697_s_at | 0.3336 | 0.0612 | |
| GSE29001 | CHGA | 1113 | 204697_s_at | -0.2285 | 0.1949 | |
| GSE38129 | CHGA | 1113 | 204697_s_at | -0.3305 | 0.1527 | |
| GSE45670 | CHGA | 1113 | 204697_s_at | 0.2596 | 0.4475 | |
| GSE53622 | CHGA | 1113 | 47435 | -0.3751 | 0.3550 | |
| GSE53624 | CHGA | 1113 | 47435 | -0.2825 | 0.3525 | |
| GSE63941 | CHGA | 1113 | 204697_s_at | 0.2318 | 0.2059 | |
| GSE77861 | CHGA | 1113 | 204697_s_at | -0.0542 | 0.7261 | |
| GSE97050 | CHGA | 1113 | A_33_P3293164 | 0.2671 | 0.3605 | |
| SRP133303 | CHGA | 1113 | RNAseq | -1.4059 | 0.0094 | |
| TCGA | CHGA | 1113 | RNAseq | -3.7738 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 2.
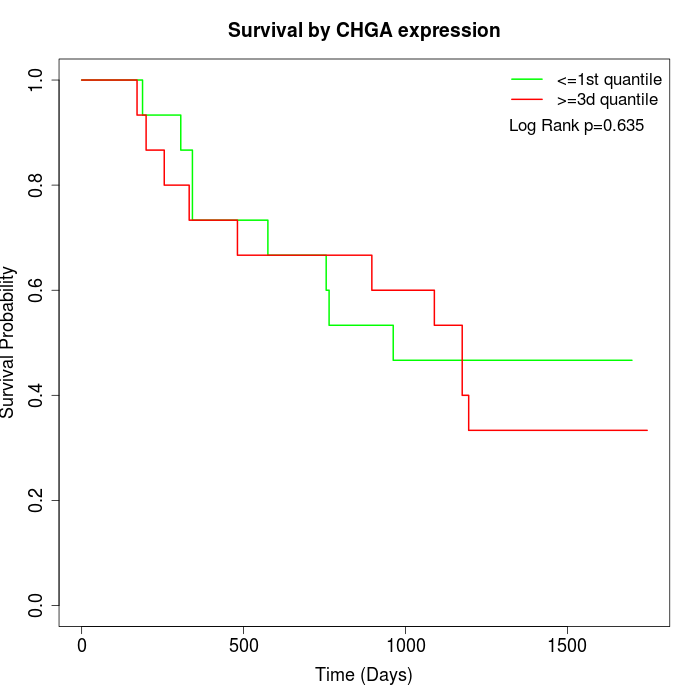
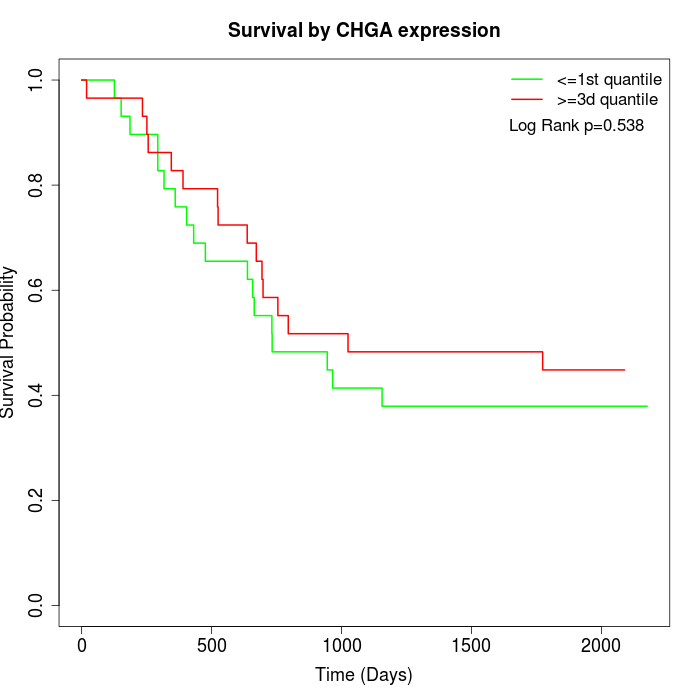
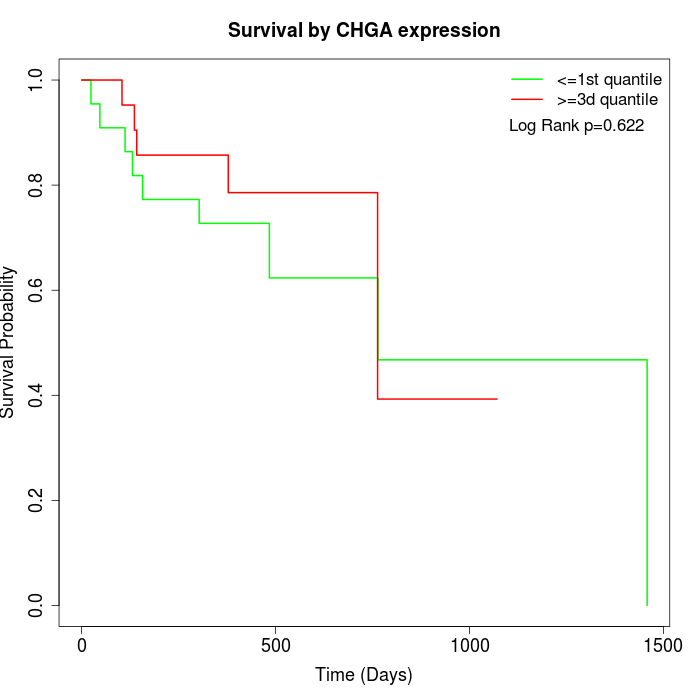
Survival by CHGA expression:
Note: Click image to view full size file.
Copy number change of CHGA:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CHGA | 1113 | 8 | 5 | 17 | |
| GSE20123 | CHGA | 1113 | 8 | 4 | 18 | |
| GSE43470 | CHGA | 1113 | 7 | 3 | 33 | |
| GSE46452 | CHGA | 1113 | 16 | 3 | 40 | |
| GSE47630 | CHGA | 1113 | 11 | 8 | 21 | |
| GSE54993 | CHGA | 1113 | 3 | 8 | 59 | |
| GSE54994 | CHGA | 1113 | 20 | 4 | 29 | |
| GSE60625 | CHGA | 1113 | 0 | 2 | 9 | |
| GSE74703 | CHGA | 1113 | 6 | 3 | 27 | |
| GSE74704 | CHGA | 1113 | 4 | 3 | 13 | |
| TCGA | CHGA | 1113 | 29 | 21 | 46 |
Total number of gains: 112; Total number of losses: 64; Total Number of normals: 312.
Somatic mutations of CHGA:
Generating mutation plots.
Highly correlated genes for CHGA:
Showing top 20/464 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CHGA | IFNA10 | 0.763702 | 3 | 0 | 3 |
| CHGA | SLC8A3 | 0.74959 | 3 | 0 | 3 |
| CHGA | KIR2DS3 | 0.723306 | 4 | 0 | 4 |
| CHGA | WNT10B | 0.715676 | 3 | 0 | 3 |
| CHGA | C14orf178 | 0.710676 | 3 | 0 | 3 |
| CHGA | MTTP | 0.70528 | 4 | 0 | 4 |
| CHGA | ZMAT4 | 0.701609 | 4 | 0 | 4 |
| CHGA | C11orf16 | 0.695446 | 3 | 0 | 3 |
| CHGA | KCNS1 | 0.694726 | 3 | 0 | 3 |
| CHGA | CDKL2 | 0.691403 | 3 | 0 | 3 |
| CHGA | PDGFB | 0.687276 | 3 | 0 | 3 |
| CHGA | DPF3 | 0.686007 | 3 | 0 | 3 |
| CHGA | CGA | 0.685296 | 3 | 0 | 3 |
| CHGA | GC | 0.683052 | 5 | 0 | 5 |
| CHGA | DPYSL5 | 0.676331 | 3 | 0 | 3 |
| CHGA | MYL3 | 0.670165 | 3 | 0 | 3 |
| CHGA | TAS2R7 | 0.66873 | 4 | 0 | 3 |
| CHGA | GKN2 | 0.66589 | 3 | 0 | 3 |
| CHGA | TULP2 | 0.66538 | 4 | 0 | 4 |
| CHGA | SERPIND1 | 0.662831 | 4 | 0 | 3 |
For details and further investigation, click here