| Full name: Fc receptor like 4 | Alias Symbol: FCRH4|IRTA1|IGFP2|CD307d | ||
| Type: protein-coding gene | Cytoband: 1q23.1 | ||
| Entrez ID: 83417 | HGNC ID: HGNC:18507 | Ensembl Gene: ENSG00000163518 | OMIM ID: 605876 |
| Drug and gene relationship at DGIdb | |||
Expression of FCRL4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | FCRL4 | 83417 | 224403_at | -0.1566 | 0.7306 | |
| GSE26886 | FCRL4 | 83417 | 224403_at | 0.1850 | 0.4937 | |
| GSE45670 | FCRL4 | 83417 | 224403_at | 0.0668 | 0.6930 | |
| GSE53622 | FCRL4 | 83417 | 35048 | -0.0305 | 0.7744 | |
| GSE53624 | FCRL4 | 83417 | 35048 | -0.6649 | 0.0000 | |
| GSE63941 | FCRL4 | 83417 | 224401_s_at | -0.0838 | 0.4595 | |
| GSE77861 | FCRL4 | 83417 | 224403_at | -0.1040 | 0.2564 | |
| GSE97050 | FCRL4 | 83417 | A_23_P115200 | 0.3921 | 0.2515 | |
| TCGA | FCRL4 | 83417 | RNAseq | 0.2712 | 0.7443 |
Upregulated datasets: 0; Downregulated datasets: 0.
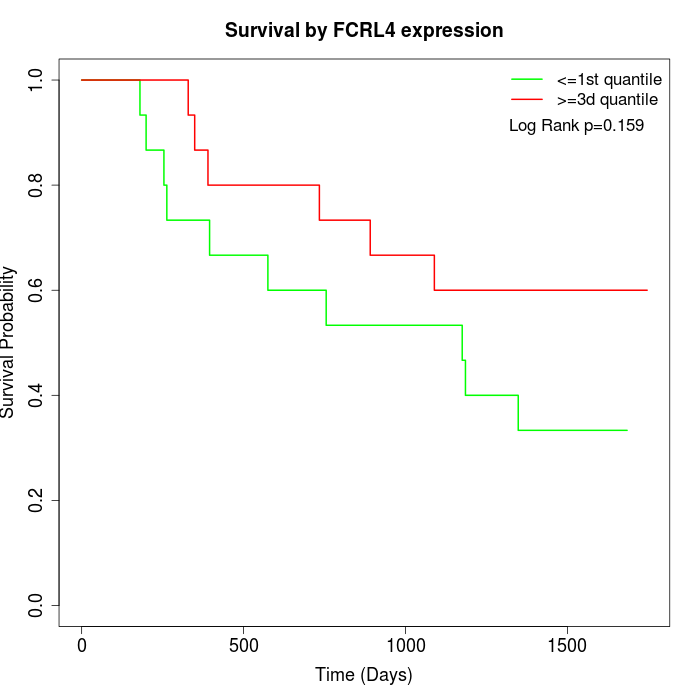
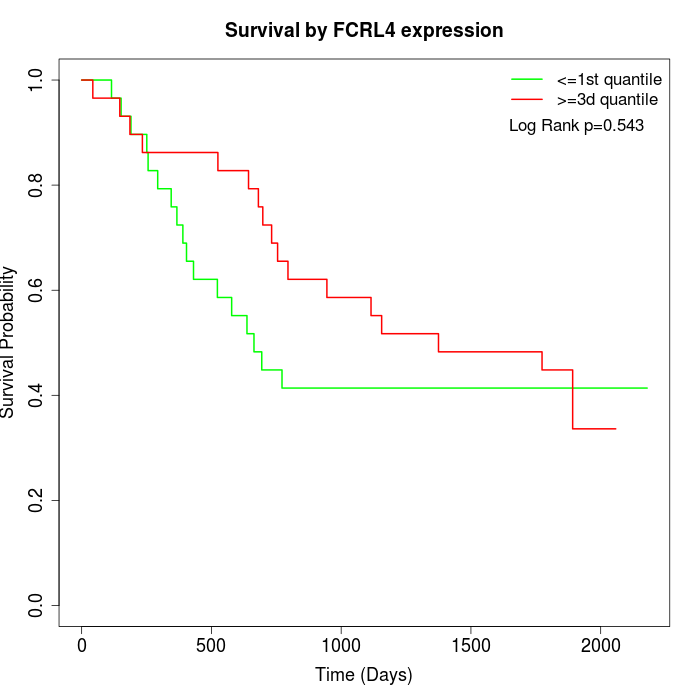
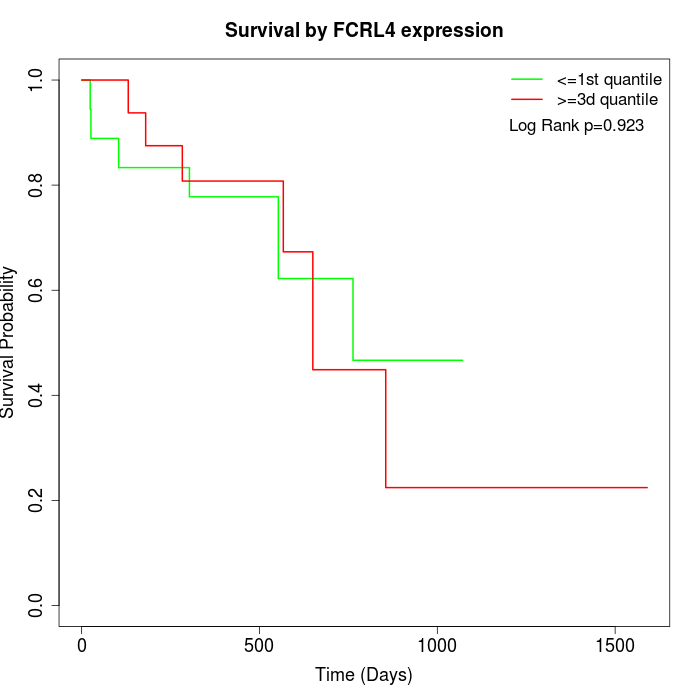
Survival by FCRL4 expression:
Note: Click image to view full size file.
Copy number change of FCRL4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | FCRL4 | 83417 | 9 | 1 | 20 | |
| GSE20123 | FCRL4 | 83417 | 9 | 0 | 21 | |
| GSE43470 | FCRL4 | 83417 | 7 | 2 | 34 | |
| GSE46452 | FCRL4 | 83417 | 2 | 1 | 56 | |
| GSE47630 | FCRL4 | 83417 | 14 | 0 | 26 | |
| GSE54993 | FCRL4 | 83417 | 0 | 5 | 65 | |
| GSE54994 | FCRL4 | 83417 | 16 | 0 | 37 | |
| GSE60625 | FCRL4 | 83417 | 0 | 0 | 11 | |
| GSE74703 | FCRL4 | 83417 | 7 | 2 | 27 | |
| GSE74704 | FCRL4 | 83417 | 4 | 0 | 16 | |
| TCGA | FCRL4 | 83417 | 39 | 2 | 55 |
Total number of gains: 107; Total number of losses: 13; Total Number of normals: 368.
Somatic mutations of FCRL4:
Generating mutation plots.
Highly correlated genes for FCRL4:
Showing top 20/86 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| FCRL4 | RARRES1 | 0.696382 | 3 | 0 | 3 |
| FCRL4 | FLT3LG | 0.694568 | 3 | 0 | 3 |
| FCRL4 | AMPD1 | 0.69452 | 3 | 0 | 3 |
| FCRL4 | EXOC3L4 | 0.682447 | 3 | 0 | 3 |
| FCRL4 | ABCB4 | 0.679697 | 4 | 0 | 4 |
| FCRL4 | CEP128 | 0.666435 | 3 | 0 | 3 |
| FCRL4 | CHRM1 | 0.665985 | 3 | 0 | 3 |
| FCRL4 | GRAP2 | 0.662117 | 3 | 0 | 3 |
| FCRL4 | FCRL3 | 0.660083 | 5 | 0 | 4 |
| FCRL4 | C16orf54 | 0.655022 | 4 | 0 | 4 |
| FCRL4 | CD19 | 0.655015 | 6 | 0 | 6 |
| FCRL4 | OGFRL1 | 0.651518 | 3 | 0 | 3 |
| FCRL4 | KCNE3 | 0.646672 | 3 | 0 | 3 |
| FCRL4 | PTPN22 | 0.642584 | 3 | 0 | 3 |
| FCRL4 | FCRLA | 0.641097 | 6 | 0 | 5 |
| FCRL4 | DOK1 | 0.635694 | 3 | 0 | 3 |
| FCRL4 | CCL28 | 0.628768 | 4 | 0 | 3 |
| FCRL4 | RAB39B | 0.623608 | 4 | 0 | 3 |
| FCRL4 | METTL6 | 0.62144 | 4 | 0 | 4 |
| FCRL4 | KCNA3 | 0.620438 | 4 | 0 | 3 |
For details and further investigation, click here