| Full name: carbohydrate sulfotransferase 2 | Alias Symbol: C6ST | ||
| Type: protein-coding gene | Cytoband: 3q24 | ||
| Entrez ID: 9435 | HGNC ID: HGNC:1970 | Ensembl Gene: ENSG00000175040 | OMIM ID: 603798 |
| Drug and gene relationship at DGIdb | |||
Expression of CHST2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CHST2 | 9435 | 203921_at | 2.3192 | 0.1214 | |
| GSE20347 | CHST2 | 9435 | 203921_at | 1.5303 | 0.0000 | |
| GSE23400 | CHST2 | 9435 | 203921_at | 1.1314 | 0.0000 | |
| GSE26886 | CHST2 | 9435 | 203921_at | 0.7751 | 0.0084 | |
| GSE29001 | CHST2 | 9435 | 203921_at | 1.7793 | 0.0002 | |
| GSE38129 | CHST2 | 9435 | 203921_at | 1.3054 | 0.0000 | |
| GSE45670 | CHST2 | 9435 | 203921_at | 1.8414 | 0.0030 | |
| GSE53622 | CHST2 | 9435 | 40492 | 1.2626 | 0.0000 | |
| GSE53624 | CHST2 | 9435 | 40492 | 1.7381 | 0.0000 | |
| GSE63941 | CHST2 | 9435 | 203921_at | -1.3805 | 0.0962 | |
| GSE77861 | CHST2 | 9435 | 203921_at | 1.1919 | 0.0138 | |
| GSE97050 | CHST2 | 9435 | A_23_P40847 | 0.6025 | 0.1406 | |
| SRP007169 | CHST2 | 9435 | RNAseq | 5.8522 | 0.0000 | |
| SRP008496 | CHST2 | 9435 | RNAseq | 4.0792 | 0.0000 | |
| SRP064894 | CHST2 | 9435 | RNAseq | 2.5970 | 0.0000 | |
| SRP133303 | CHST2 | 9435 | RNAseq | 1.5949 | 0.0000 | |
| SRP159526 | CHST2 | 9435 | RNAseq | 3.4961 | 0.0000 | |
| SRP193095 | CHST2 | 9435 | RNAseq | 2.7157 | 0.0000 | |
| SRP219564 | CHST2 | 9435 | RNAseq | 1.0180 | 0.1134 | |
| TCGA | CHST2 | 9435 | RNAseq | 0.6049 | 0.0004 |
Upregulated datasets: 14; Downregulated datasets: 0.
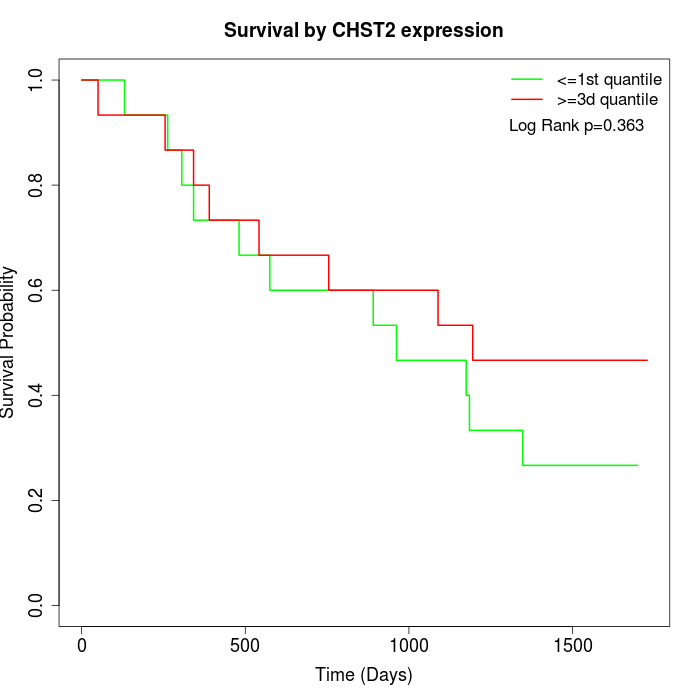
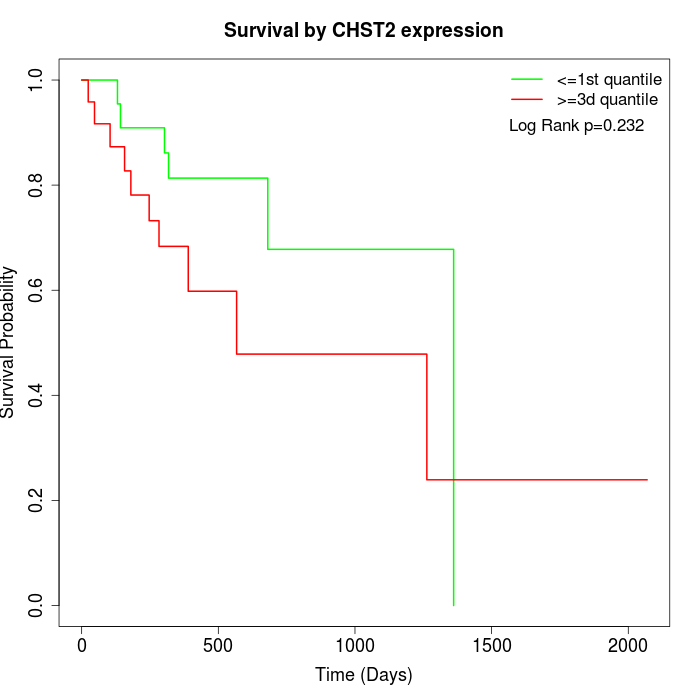
Survival by CHST2 expression:
Note: Click image to view full size file.
Copy number change of CHST2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CHST2 | 9435 | 20 | 0 | 10 | |
| GSE20123 | CHST2 | 9435 | 20 | 0 | 10 | |
| GSE43470 | CHST2 | 9435 | 22 | 0 | 21 | |
| GSE46452 | CHST2 | 9435 | 15 | 4 | 40 | |
| GSE47630 | CHST2 | 9435 | 19 | 3 | 18 | |
| GSE54993 | CHST2 | 9435 | 1 | 8 | 61 | |
| GSE54994 | CHST2 | 9435 | 34 | 1 | 18 | |
| GSE60625 | CHST2 | 9435 | 0 | 6 | 5 | |
| GSE74703 | CHST2 | 9435 | 19 | 0 | 17 | |
| GSE74704 | CHST2 | 9435 | 14 | 0 | 6 | |
| TCGA | CHST2 | 9435 | 64 | 3 | 29 |
Total number of gains: 228; Total number of losses: 25; Total Number of normals: 235.
Somatic mutations of CHST2:
Generating mutation plots.
Highly correlated genes for CHST2:
Showing top 20/1357 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CHST2 | NEIL2 | 0.780316 | 3 | 0 | 3 |
| CHST2 | FAM89A | 0.76094 | 3 | 0 | 3 |
| CHST2 | ZC3H7B | 0.758691 | 3 | 0 | 3 |
| CHST2 | TPCN2 | 0.735619 | 4 | 0 | 3 |
| CHST2 | C1orf226 | 0.726492 | 5 | 0 | 4 |
| CHST2 | YWHAG | 0.726115 | 4 | 0 | 4 |
| CHST2 | C6orf141 | 0.724274 | 3 | 0 | 3 |
| CHST2 | ZBTB34 | 0.719793 | 3 | 0 | 3 |
| CHST2 | SPNS1 | 0.718975 | 4 | 0 | 4 |
| CHST2 | ALPK2 | 0.718951 | 3 | 0 | 3 |
| CHST2 | MAPK6 | 0.713608 | 4 | 0 | 3 |
| CHST2 | TRIM35 | 0.710676 | 3 | 0 | 3 |
| CHST2 | ARL13B | 0.710248 | 6 | 0 | 5 |
| CHST2 | SPRY4-IT1 | 0.709934 | 3 | 0 | 3 |
| CHST2 | EGLN2 | 0.703091 | 3 | 0 | 3 |
| CHST2 | LRP12 | 0.702943 | 12 | 0 | 12 |
| CHST2 | GLIS3 | 0.695443 | 4 | 0 | 4 |
| CHST2 | AGPAT4 | 0.695025 | 4 | 0 | 4 |
| CHST2 | LIX1L | 0.694474 | 3 | 0 | 3 |
| CHST2 | GSPT2 | 0.693311 | 4 | 0 | 3 |
For details and further investigation, click here