| Full name: cytoplasmic linker associated protein 2 | Alias Symbol: KIAA0627 | ||
| Type: protein-coding gene | Cytoband: 3p22.3 | ||
| Entrez ID: 23122 | HGNC ID: HGNC:17078 | Ensembl Gene: ENSG00000163539 | OMIM ID: 605853 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of CLASP2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CLASP2 | 23122 | 212306_at | 0.0169 | 0.9813 | |
| GSE20347 | CLASP2 | 23122 | 212306_at | -0.2926 | 0.0623 | |
| GSE23400 | CLASP2 | 23122 | 212306_at | 0.0436 | 0.5113 | |
| GSE26886 | CLASP2 | 23122 | 212306_at | -0.2227 | 0.3354 | |
| GSE29001 | CLASP2 | 23122 | 212309_at | -0.2272 | 0.2289 | |
| GSE38129 | CLASP2 | 23122 | 212306_at | -0.3095 | 0.0108 | |
| GSE45670 | CLASP2 | 23122 | 212306_at | -0.0385 | 0.8076 | |
| GSE53622 | CLASP2 | 23122 | 77511 | -0.3420 | 0.0000 | |
| GSE53624 | CLASP2 | 23122 | 19249 | -0.4176 | 0.0000 | |
| GSE63941 | CLASP2 | 23122 | 212306_at | 0.1023 | 0.8490 | |
| GSE77861 | CLASP2 | 23122 | 212306_at | -0.1333 | 0.5800 | |
| GSE97050 | CLASP2 | 23122 | A_23_P44295 | -0.5418 | 0.1889 | |
| SRP007169 | CLASP2 | 23122 | RNAseq | 0.0777 | 0.7993 | |
| SRP008496 | CLASP2 | 23122 | RNAseq | 0.1216 | 0.5026 | |
| SRP064894 | CLASP2 | 23122 | RNAseq | -0.3666 | 0.0773 | |
| SRP133303 | CLASP2 | 23122 | RNAseq | -0.3875 | 0.0013 | |
| SRP159526 | CLASP2 | 23122 | RNAseq | -0.5840 | 0.0032 | |
| SRP193095 | CLASP2 | 23122 | RNAseq | -0.3647 | 0.0000 | |
| SRP219564 | CLASP2 | 23122 | RNAseq | -0.3625 | 0.2117 | |
| TCGA | CLASP2 | 23122 | RNAseq | -0.1704 | 0.0005 |
Upregulated datasets: 0; Downregulated datasets: 0.
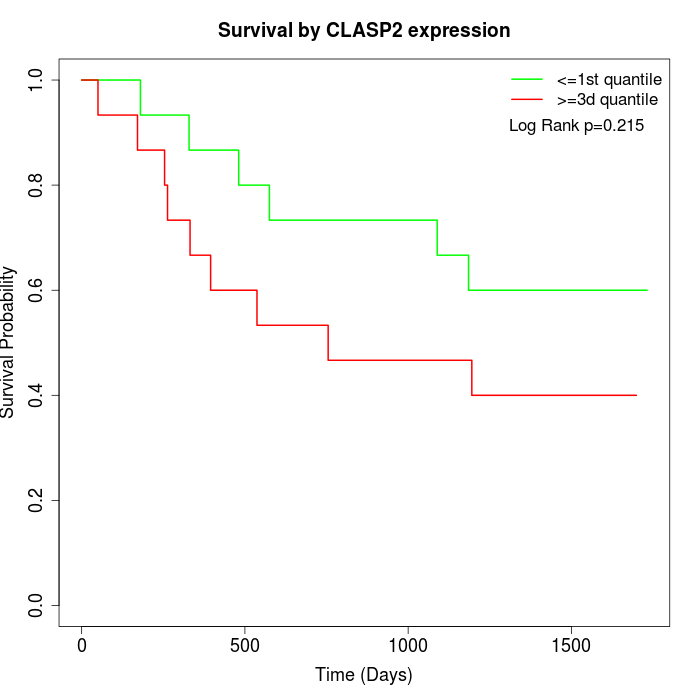
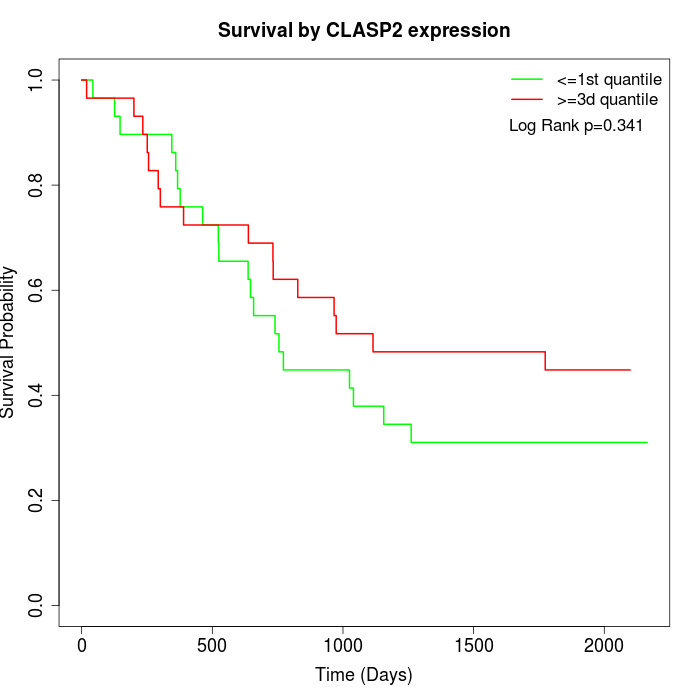
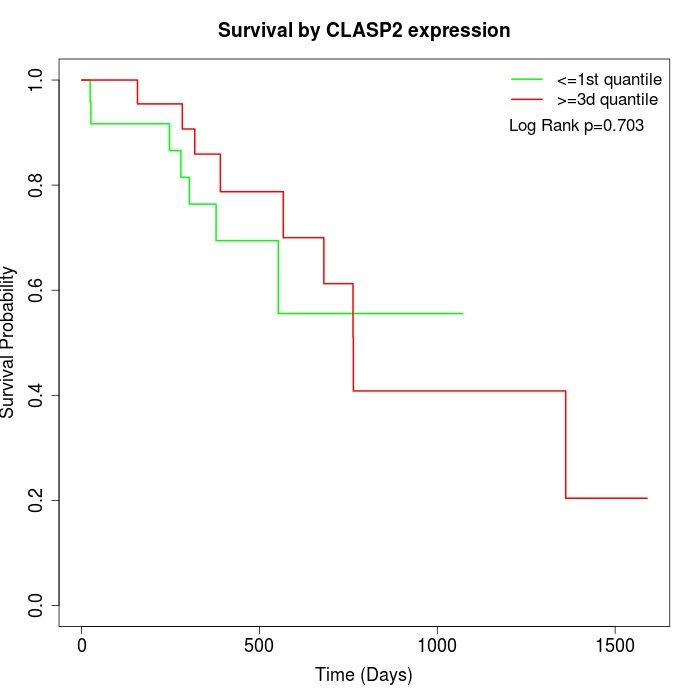
Survival by CLASP2 expression:
Note: Click image to view full size file.
Copy number change of CLASP2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CLASP2 | 23122 | 0 | 18 | 12 | |
| GSE20123 | CLASP2 | 23122 | 0 | 18 | 12 | |
| GSE43470 | CLASP2 | 23122 | 1 | 19 | 23 | |
| GSE46452 | CLASP2 | 23122 | 2 | 17 | 40 | |
| GSE47630 | CLASP2 | 23122 | 1 | 25 | 14 | |
| GSE54993 | CLASP2 | 23122 | 6 | 3 | 61 | |
| GSE54994 | CLASP2 | 23122 | 1 | 32 | 20 | |
| GSE60625 | CLASP2 | 23122 | 5 | 0 | 6 | |
| GSE74703 | CLASP2 | 23122 | 1 | 16 | 19 | |
| GSE74704 | CLASP2 | 23122 | 0 | 12 | 8 | |
| TCGA | CLASP2 | 23122 | 1 | 69 | 26 |
Total number of gains: 18; Total number of losses: 229; Total Number of normals: 241.
Somatic mutations of CLASP2:
Generating mutation plots.
Highly correlated genes for CLASP2:
Showing top 20/248 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CLASP2 | CCDC51 | 0.777331 | 3 | 0 | 3 |
| CLASP2 | TUBB2A | 0.725287 | 3 | 0 | 3 |
| CLASP2 | SRFBP1 | 0.718624 | 3 | 0 | 3 |
| CLASP2 | PYGM | 0.695482 | 3 | 0 | 3 |
| CLASP2 | SLMAP | 0.692739 | 4 | 0 | 4 |
| CLASP2 | RAB9B | 0.688491 | 3 | 0 | 3 |
| CLASP2 | FNDC5 | 0.683675 | 3 | 0 | 3 |
| CLASP2 | RCAN2 | 0.683562 | 3 | 0 | 3 |
| CLASP2 | TNRC6A | 0.682232 | 3 | 0 | 3 |
| CLASP2 | ZCCHC10 | 0.680683 | 3 | 0 | 3 |
| CLASP2 | CASQ2 | 0.679116 | 3 | 0 | 3 |
| CLASP2 | ANGPTL1 | 0.678929 | 3 | 0 | 3 |
| CLASP2 | AGPAT5 | 0.675052 | 3 | 0 | 3 |
| CLASP2 | ANO5 | 0.674491 | 3 | 0 | 3 |
| CLASP2 | SUPT7L | 0.672351 | 3 | 0 | 3 |
| CLASP2 | NEGR1 | 0.670999 | 3 | 0 | 3 |
| CLASP2 | ATG2B | 0.670474 | 4 | 0 | 4 |
| CLASP2 | SEC23A | 0.669725 | 4 | 0 | 4 |
| CLASP2 | ROCK2 | 0.660978 | 4 | 0 | 3 |
| CLASP2 | USP24 | 0.652511 | 3 | 0 | 3 |
For details and further investigation, click here