| Full name: claudin domain containing 2 | Alias Symbol: MGC33839 | ||
| Type: protein-coding gene | Cytoband: 19q13.41 | ||
| Entrez ID: 125875 | HGNC ID: HGNC:28511 | Ensembl Gene: ENSG00000160318 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of CLDND2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CLDND2 | 125875 | 231162_at | -0.3010 | 0.4709 | |
| GSE26886 | CLDND2 | 125875 | 231162_at | 0.2996 | 0.0159 | |
| GSE45670 | CLDND2 | 125875 | 231162_at | -0.1222 | 0.2054 | |
| GSE53622 | CLDND2 | 125875 | 57460 | 0.0173 | 0.9293 | |
| GSE53624 | CLDND2 | 125875 | 57460 | -0.1057 | 0.4154 | |
| GSE63941 | CLDND2 | 125875 | 231162_at | -0.1518 | 0.4870 | |
| GSE77861 | CLDND2 | 125875 | 231162_at | -0.2579 | 0.0390 | |
| GSE97050 | CLDND2 | 125875 | A_23_P374902 | -0.0588 | 0.8389 | |
| SRP133303 | CLDND2 | 125875 | RNAseq | -0.3980 | 0.1783 | |
| SRP159526 | CLDND2 | 125875 | RNAseq | 0.1550 | 0.7851 | |
| SRP219564 | CLDND2 | 125875 | RNAseq | 0.6046 | 0.3157 | |
| TCGA | CLDND2 | 125875 | RNAseq | -0.5705 | 0.2256 |
Upregulated datasets: 0; Downregulated datasets: 0.
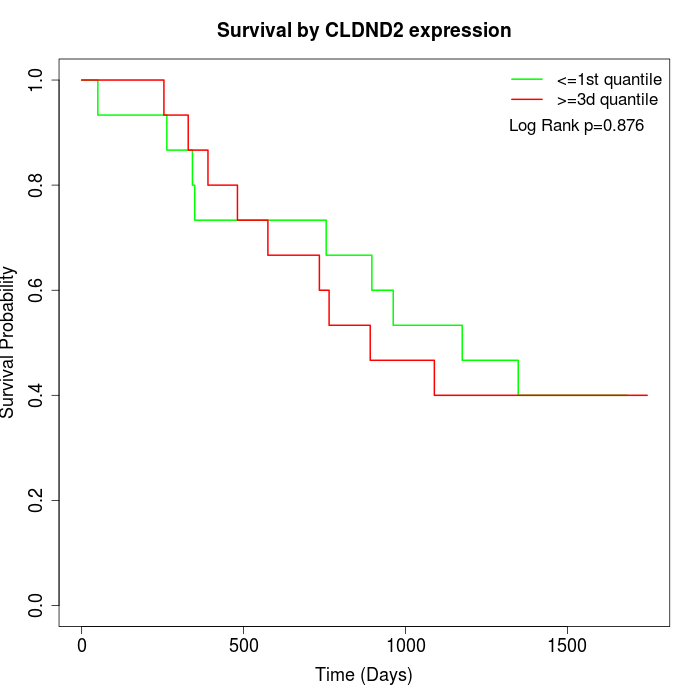
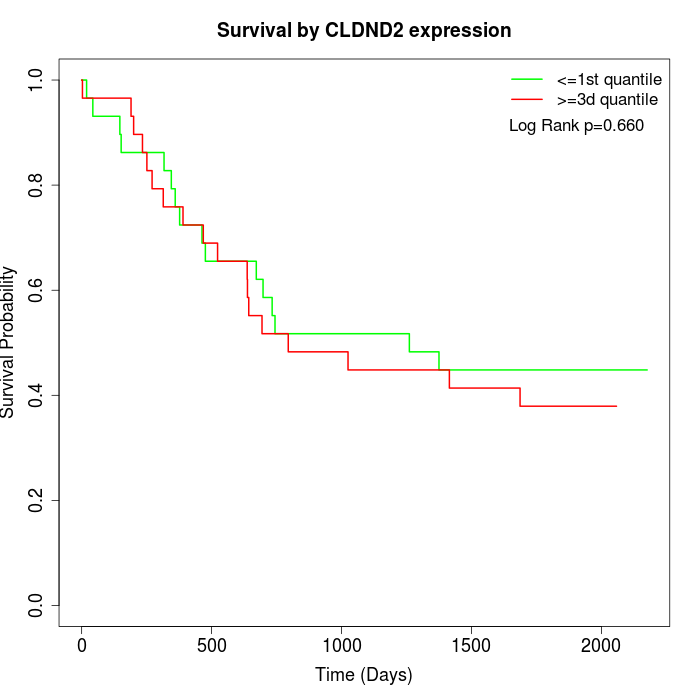
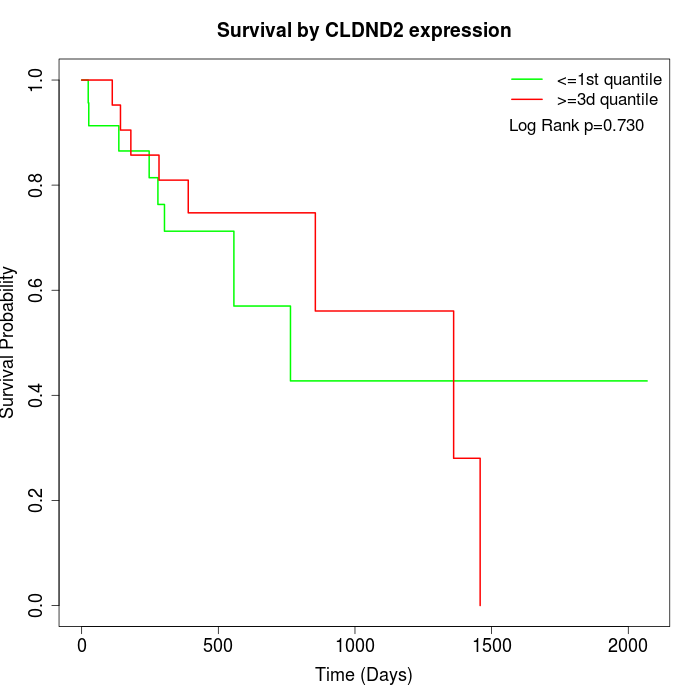
Survival by CLDND2 expression:
Note: Click image to view full size file.
Copy number change of CLDND2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CLDND2 | 125875 | 3 | 4 | 23 | |
| GSE20123 | CLDND2 | 125875 | 3 | 3 | 24 | |
| GSE43470 | CLDND2 | 125875 | 3 | 12 | 28 | |
| GSE46452 | CLDND2 | 125875 | 45 | 1 | 13 | |
| GSE47630 | CLDND2 | 125875 | 9 | 6 | 25 | |
| GSE54993 | CLDND2 | 125875 | 17 | 4 | 49 | |
| GSE54994 | CLDND2 | 125875 | 4 | 14 | 35 | |
| GSE60625 | CLDND2 | 125875 | 9 | 0 | 2 | |
| GSE74703 | CLDND2 | 125875 | 3 | 8 | 25 | |
| GSE74704 | CLDND2 | 125875 | 3 | 1 | 16 | |
| TCGA | CLDND2 | 125875 | 14 | 19 | 63 |
Total number of gains: 113; Total number of losses: 72; Total Number of normals: 303.
Somatic mutations of CLDND2:
Generating mutation plots.
Highly correlated genes for CLDND2:
Showing top 20/184 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CLDND2 | C22orf31 | 0.828357 | 3 | 0 | 3 |
| CLDND2 | CLDN2 | 0.795801 | 3 | 0 | 3 |
| CLDND2 | GRAP2 | 0.794085 | 3 | 0 | 3 |
| CLDND2 | HEYL | 0.785351 | 3 | 0 | 3 |
| CLDND2 | EIF3L | 0.768483 | 3 | 0 | 3 |
| CLDND2 | AVP | 0.765301 | 3 | 0 | 3 |
| CLDND2 | SMTNL2 | 0.762057 | 3 | 0 | 3 |
| CLDND2 | USP9Y | 0.7604 | 3 | 0 | 3 |
| CLDND2 | GNGT2 | 0.755097 | 3 | 0 | 3 |
| CLDND2 | C11orf96 | 0.75221 | 3 | 0 | 3 |
| CLDND2 | GYPC | 0.733212 | 3 | 0 | 3 |
| CLDND2 | KLHL17 | 0.731042 | 3 | 0 | 3 |
| CLDND2 | SYNE1 | 0.729728 | 3 | 0 | 3 |
| CLDND2 | SH3BP5 | 0.72624 | 3 | 0 | 3 |
| CLDND2 | ATOX1 | 0.725001 | 3 | 0 | 3 |
| CLDND2 | EVC2 | 0.719892 | 4 | 0 | 4 |
| CLDND2 | PCGF2 | 0.718401 | 3 | 0 | 3 |
| CLDND2 | RBP2 | 0.716321 | 3 | 0 | 3 |
| CLDND2 | VAMP2 | 0.715484 | 3 | 0 | 3 |
| CLDND2 | ATP8B2 | 0.71272 | 4 | 0 | 4 |
For details and further investigation, click here