| Full name: clathrin heavy chain | Alias Symbol: Hc | ||
| Type: protein-coding gene | Cytoband: 17q23.1 | ||
| Entrez ID: 1213 | HGNC ID: HGNC:2092 | Ensembl Gene: ENSG00000141367 | OMIM ID: 118955 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
CLTC involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa05016 | Huntington's disease |
Expression of CLTC:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CLTC | 1213 | 200614_at | 0.3093 | 0.2643 | |
| GSE20347 | CLTC | 1213 | 200614_at | 0.0908 | 0.4406 | |
| GSE23400 | CLTC | 1213 | 200614_at | 0.2008 | 0.0000 | |
| GSE26886 | CLTC | 1213 | 200614_at | 0.1610 | 0.2864 | |
| GSE29001 | CLTC | 1213 | 200614_at | -0.0239 | 0.9216 | |
| GSE38129 | CLTC | 1213 | 200614_at | 0.2031 | 0.0092 | |
| GSE45670 | CLTC | 1213 | 200614_at | 0.1640 | 0.1116 | |
| GSE53622 | CLTC | 1213 | 62910 | -0.1065 | 0.1461 | |
| GSE53624 | CLTC | 1213 | 60246 | 0.2728 | 0.0004 | |
| GSE63941 | CLTC | 1213 | 200614_at | -0.4346 | 0.1575 | |
| GSE77861 | CLTC | 1213 | 200614_at | 0.5436 | 0.0086 | |
| GSE97050 | CLTC | 1213 | A_33_P3274731 | -0.1605 | 0.5673 | |
| SRP007169 | CLTC | 1213 | RNAseq | 0.1091 | 0.6866 | |
| SRP008496 | CLTC | 1213 | RNAseq | 0.0967 | 0.5198 | |
| SRP064894 | CLTC | 1213 | RNAseq | 0.1706 | 0.2776 | |
| SRP133303 | CLTC | 1213 | RNAseq | 0.5765 | 0.0002 | |
| SRP159526 | CLTC | 1213 | RNAseq | 0.1172 | 0.4829 | |
| SRP193095 | CLTC | 1213 | RNAseq | 0.0310 | 0.7786 | |
| SRP219564 | CLTC | 1213 | RNAseq | -0.0248 | 0.9386 | |
| TCGA | CLTC | 1213 | RNAseq | -0.0077 | 0.8327 |
Upregulated datasets: 0; Downregulated datasets: 0.
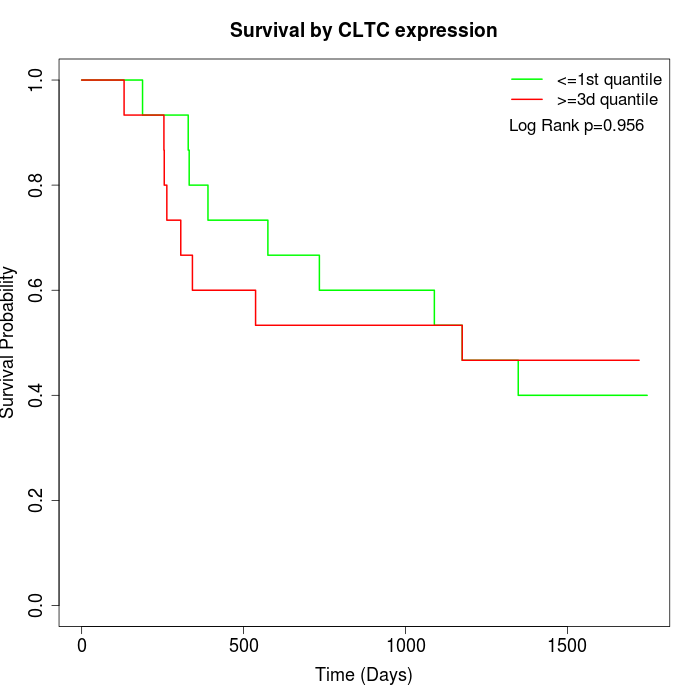
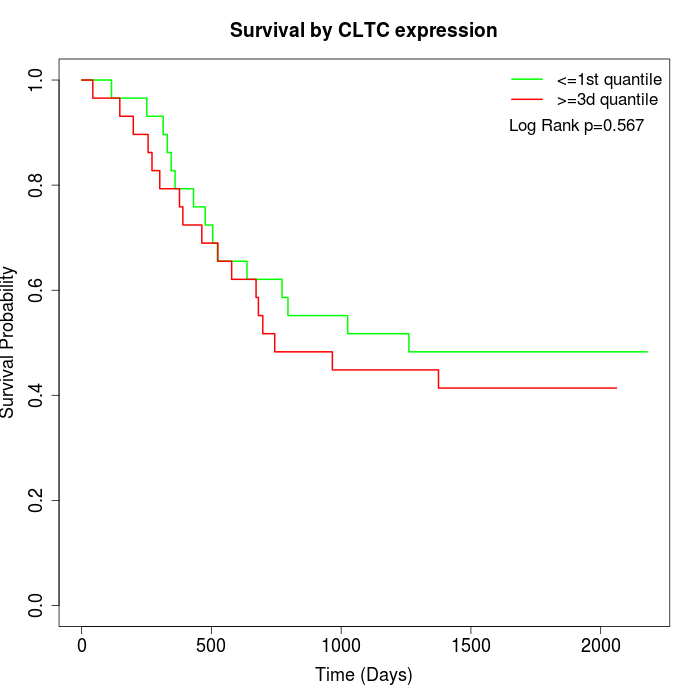
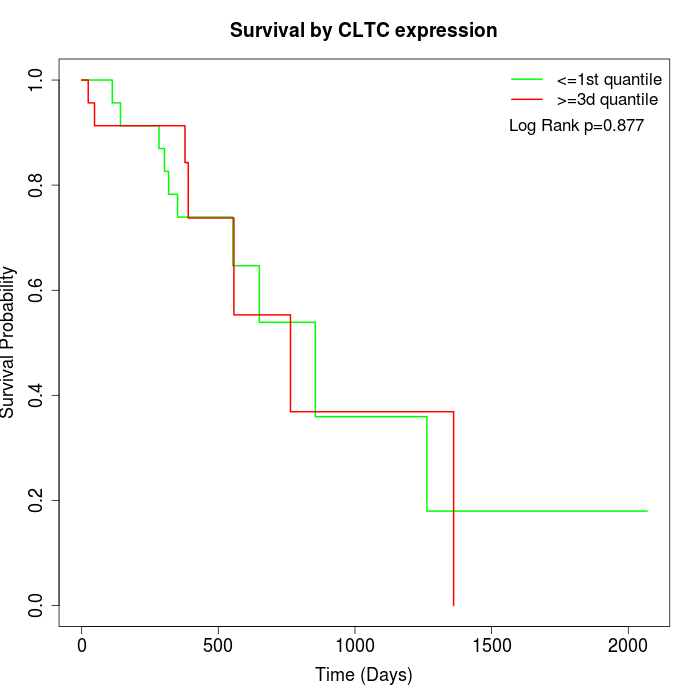
Survival by CLTC expression:
Note: Click image to view full size file.
Copy number change of CLTC:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CLTC | 1213 | 6 | 1 | 23 | |
| GSE20123 | CLTC | 1213 | 6 | 1 | 23 | |
| GSE43470 | CLTC | 1213 | 5 | 0 | 38 | |
| GSE46452 | CLTC | 1213 | 32 | 0 | 27 | |
| GSE47630 | CLTC | 1213 | 7 | 1 | 32 | |
| GSE54993 | CLTC | 1213 | 2 | 5 | 63 | |
| GSE54994 | CLTC | 1213 | 9 | 5 | 39 | |
| GSE60625 | CLTC | 1213 | 4 | 0 | 7 | |
| GSE74703 | CLTC | 1213 | 5 | 0 | 31 | |
| GSE74704 | CLTC | 1213 | 5 | 1 | 14 | |
| TCGA | CLTC | 1213 | 26 | 8 | 62 |
Total number of gains: 107; Total number of losses: 22; Total Number of normals: 359.
Somatic mutations of CLTC:
Generating mutation plots.
Highly correlated genes for CLTC:
Showing top 20/63 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CLTC | CCL26 | 0.660812 | 4 | 0 | 4 |
| CLTC | TCEAL7 | 0.655615 | 3 | 0 | 3 |
| CLTC | PRAME | 0.637203 | 6 | 0 | 5 |
| CLTC | PSMD11 | 0.623549 | 8 | 0 | 6 |
| CLTC | TMEM9 | 0.612082 | 4 | 0 | 4 |
| CLTC | RANBP1 | 0.611472 | 7 | 0 | 6 |
| CLTC | COPRS | 0.607079 | 3 | 0 | 3 |
| CLTC | CCDC43 | 0.599334 | 5 | 0 | 3 |
| CLTC | TBX3 | 0.588973 | 7 | 0 | 4 |
| CLTC | CDK17 | 0.587268 | 6 | 0 | 3 |
| CLTC | ZBTB24 | 0.586685 | 6 | 0 | 5 |
| CLTC | GREM1 | 0.582101 | 3 | 0 | 3 |
| CLTC | MAD1L1 | 0.578833 | 5 | 0 | 4 |
| CLTC | FAM131A | 0.577535 | 5 | 0 | 3 |
| CLTC | POR | 0.575038 | 5 | 0 | 3 |
| CLTC | TANGO2 | 0.574884 | 5 | 0 | 4 |
| CLTC | INTS2 | 0.566277 | 5 | 0 | 4 |
| CLTC | ERC2 | 0.56432 | 4 | 0 | 3 |
| CLTC | NAV3 | 0.563589 | 5 | 0 | 3 |
| CLTC | PSMB5 | 0.561556 | 8 | 0 | 6 |
For details and further investigation, click here