| Full name: complement C3b/C4b receptor 1 like | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 1q32.2 | ||
| Entrez ID: 1379 | HGNC ID: HGNC:2335 | Ensembl Gene: ENSG00000197721 | OMIM ID: 605886 |
| Drug and gene relationship at DGIdb | |||
Expression of CR1L:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CR1L | 1379 | 239206_at | -0.0021 | 0.9948 | |
| GSE26886 | CR1L | 1379 | 239206_at | 0.0033 | 0.9797 | |
| GSE45670 | CR1L | 1379 | 239206_at | 0.0329 | 0.7249 | |
| GSE53622 | CR1L | 1379 | 39846 | -0.5125 | 0.0027 | |
| GSE53624 | CR1L | 1379 | 39846 | -0.2404 | 0.0416 | |
| GSE63941 | CR1L | 1379 | 239206_at | 0.0168 | 0.9249 | |
| GSE77861 | CR1L | 1379 | 239206_at | -0.2607 | 0.0364 | |
| GSE97050 | CR1L | 1379 | A_33_P3319126 | 1.3625 | 0.1189 | |
| TCGA | CR1L | 1379 | RNAseq | 2.1369 | 0.0661 |
Upregulated datasets: 0; Downregulated datasets: 0.
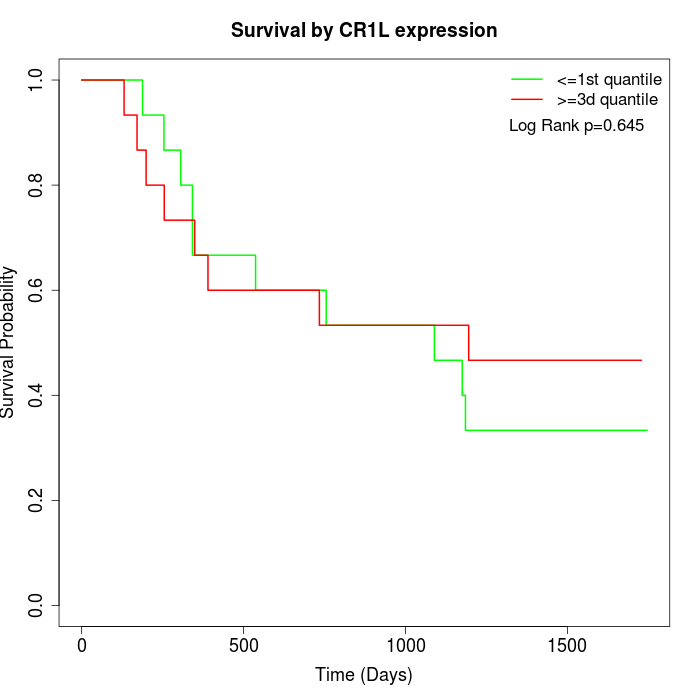
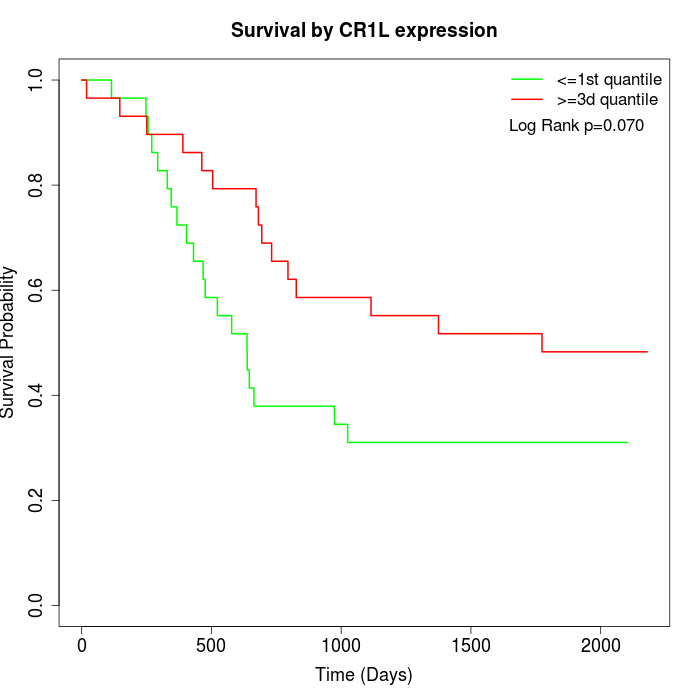
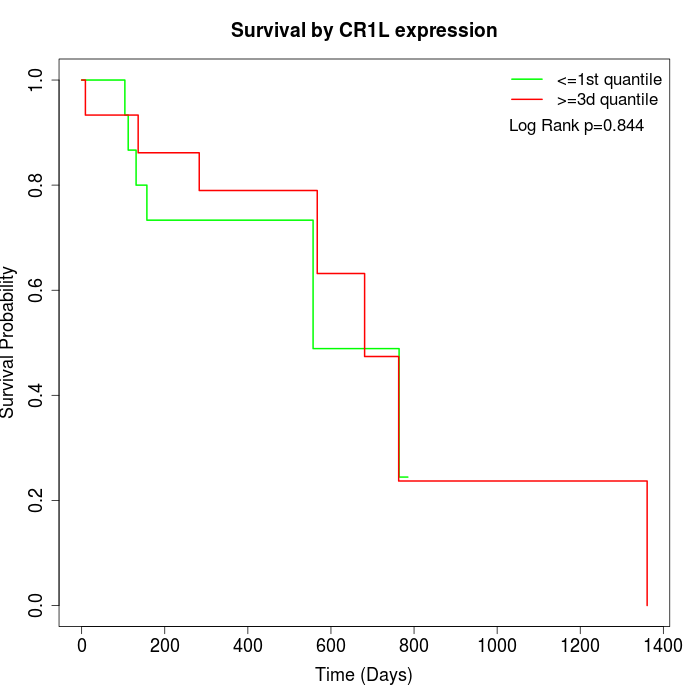
Survival by CR1L expression:
Note: Click image to view full size file.
Copy number change of CR1L:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CR1L | 1379 | 11 | 0 | 19 | |
| GSE20123 | CR1L | 1379 | 11 | 0 | 19 | |
| GSE43470 | CR1L | 1379 | 9 | 0 | 34 | |
| GSE46452 | CR1L | 1379 | 3 | 1 | 55 | |
| GSE47630 | CR1L | 1379 | 14 | 0 | 26 | |
| GSE54993 | CR1L | 1379 | 0 | 6 | 64 | |
| GSE54994 | CR1L | 1379 | 14 | 0 | 39 | |
| GSE60625 | CR1L | 1379 | 0 | 0 | 11 | |
| GSE74703 | CR1L | 1379 | 9 | 0 | 27 | |
| GSE74704 | CR1L | 1379 | 5 | 0 | 15 | |
| TCGA | CR1L | 1379 | 45 | 3 | 48 |
Total number of gains: 121; Total number of losses: 10; Total Number of normals: 357.
Somatic mutations of CR1L:
Generating mutation plots.
Highly correlated genes for CR1L:
Showing top 20/166 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CR1L | ANGPTL6 | 0.772367 | 3 | 0 | 3 |
| CR1L | PTPN22 | 0.758582 | 3 | 0 | 3 |
| CR1L | APBB1IP | 0.749164 | 4 | 0 | 4 |
| CR1L | TNFSF8 | 0.74258 | 4 | 0 | 4 |
| CR1L | PVRIG | 0.739532 | 3 | 0 | 3 |
| CR1L | BIN2 | 0.734151 | 4 | 0 | 4 |
| CR1L | ARHGAP9 | 0.732056 | 4 | 0 | 4 |
| CR1L | ATG13 | 0.725135 | 3 | 0 | 3 |
| CR1L | STAB1 | 0.723647 | 4 | 0 | 3 |
| CR1L | MAMDC4 | 0.721898 | 4 | 0 | 4 |
| CR1L | CD37 | 0.720343 | 4 | 0 | 4 |
| CR1L | IKZF1 | 0.720145 | 4 | 0 | 4 |
| CR1L | NDUFB3 | 0.695242 | 3 | 0 | 3 |
| CR1L | LILRA4 | 0.692276 | 3 | 0 | 3 |
| CR1L | DPEP2 | 0.691673 | 4 | 0 | 3 |
| CR1L | RAB27A | 0.687797 | 3 | 0 | 3 |
| CR1L | PLCB2 | 0.686322 | 4 | 0 | 3 |
| CR1L | APOL3 | 0.686164 | 3 | 0 | 3 |
| CR1L | SH3BP5 | 0.684715 | 3 | 0 | 3 |
| CR1L | CD74 | 0.683154 | 5 | 0 | 5 |
For details and further investigation, click here