| Full name: cryptochrome circadian regulator 1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 12q23.3 | ||
| Entrez ID: 1407 | HGNC ID: HGNC:2384 | Ensembl Gene: ENSG00000008405 | OMIM ID: 601933 |
| Drug and gene relationship at DGIdb | |||
CRY1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04710 | Circadian rhythm |
Expression of CRY1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CRY1 | 1407 | 209674_at | -0.7045 | 0.3377 | |
| GSE20347 | CRY1 | 1407 | 209674_at | -0.6613 | 0.0021 | |
| GSE23400 | CRY1 | 1407 | 209674_at | -0.3117 | 0.0003 | |
| GSE26886 | CRY1 | 1407 | 209674_at | 0.1150 | 0.7326 | |
| GSE29001 | CRY1 | 1407 | 209674_at | -0.4550 | 0.1851 | |
| GSE38129 | CRY1 | 1407 | 209674_at | -0.4693 | 0.0066 | |
| GSE45670 | CRY1 | 1407 | 209674_at | -0.6937 | 0.0001 | |
| GSE53622 | CRY1 | 1407 | 74504 | -0.4938 | 0.0015 | |
| GSE53624 | CRY1 | 1407 | 74504 | -0.4553 | 0.0017 | |
| GSE63941 | CRY1 | 1407 | 209674_at | -0.1957 | 0.7442 | |
| GSE77861 | CRY1 | 1407 | 209674_at | -0.2737 | 0.5769 | |
| SRP007169 | CRY1 | 1407 | RNAseq | -1.0045 | 0.0397 | |
| SRP008496 | CRY1 | 1407 | RNAseq | -0.6453 | 0.0520 | |
| SRP064894 | CRY1 | 1407 | RNAseq | -0.5151 | 0.0851 | |
| SRP133303 | CRY1 | 1407 | RNAseq | -0.2078 | 0.3516 | |
| SRP159526 | CRY1 | 1407 | RNAseq | -0.3744 | 0.3847 | |
| SRP193095 | CRY1 | 1407 | RNAseq | -0.5722 | 0.0000 | |
| SRP219564 | CRY1 | 1407 | RNAseq | -0.6001 | 0.0609 | |
| TCGA | CRY1 | 1407 | RNAseq | -0.2941 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 1.
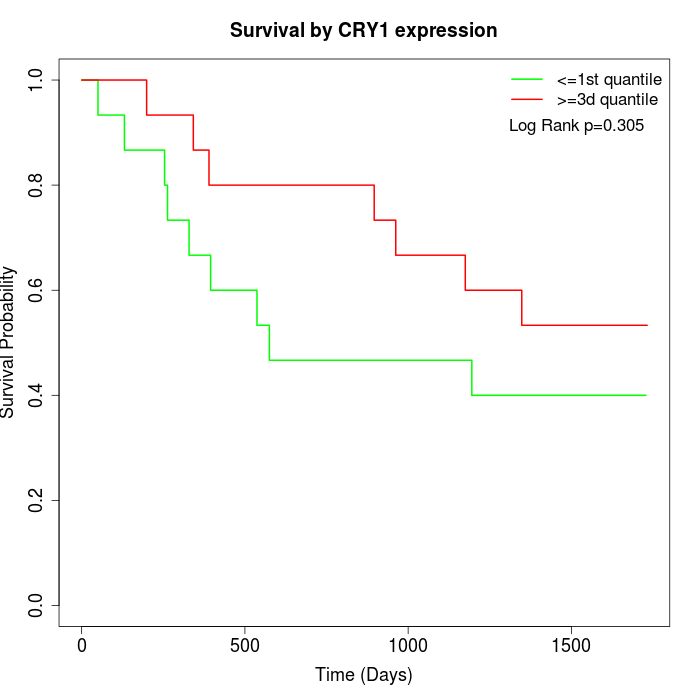
Survival by CRY1 expression:
Note: Click image to view full size file.
Copy number change of CRY1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CRY1 | 1407 | 5 | 3 | 22 | |
| GSE20123 | CRY1 | 1407 | 5 | 3 | 22 | |
| GSE43470 | CRY1 | 1407 | 2 | 1 | 40 | |
| GSE46452 | CRY1 | 1407 | 9 | 1 | 49 | |
| GSE47630 | CRY1 | 1407 | 9 | 1 | 30 | |
| GSE54993 | CRY1 | 1407 | 0 | 5 | 65 | |
| GSE54994 | CRY1 | 1407 | 4 | 2 | 47 | |
| GSE60625 | CRY1 | 1407 | 0 | 0 | 11 | |
| GSE74703 | CRY1 | 1407 | 2 | 0 | 34 | |
| GSE74704 | CRY1 | 1407 | 2 | 2 | 16 | |
| TCGA | CRY1 | 1407 | 21 | 10 | 65 |
Total number of gains: 59; Total number of losses: 28; Total Number of normals: 401.
Somatic mutations of CRY1:
Generating mutation plots.
Highly correlated genes for CRY1:
Showing top 20/118 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CRY1 | TRIP4 | 0.635235 | 4 | 0 | 3 |
| CRY1 | UCHL5 | 0.620256 | 3 | 0 | 3 |
| CRY1 | CGRRF1 | 0.60748 | 8 | 0 | 5 |
| CRY1 | EIF3J | 0.605286 | 4 | 0 | 3 |
| CRY1 | ZNF823 | 0.601707 | 3 | 0 | 3 |
| CRY1 | HERPUD2 | 0.601107 | 4 | 0 | 4 |
| CRY1 | PPP1R2 | 0.600754 | 5 | 0 | 3 |
| CRY1 | NDUFB7 | 0.60057 | 3 | 0 | 3 |
| CRY1 | ARL8B | 0.596014 | 4 | 0 | 3 |
| CRY1 | SREK1 | 0.588664 | 4 | 0 | 3 |
| CRY1 | NDUFS1 | 0.58509 | 4 | 0 | 3 |
| CRY1 | SDHD | 0.581277 | 4 | 0 | 3 |
| CRY1 | CCSER2 | 0.578949 | 6 | 0 | 5 |
| CRY1 | BECN1 | 0.577464 | 4 | 0 | 3 |
| CRY1 | MRPS31 | 0.577151 | 5 | 0 | 4 |
| CRY1 | MPP5 | 0.576291 | 4 | 0 | 3 |
| CRY1 | TBC1D12 | 0.574993 | 7 | 0 | 5 |
| CRY1 | CCNC | 0.574815 | 7 | 0 | 3 |
| CRY1 | DHRS7B | 0.57366 | 3 | 0 | 3 |
| CRY1 | UEVLD | 0.573244 | 5 | 0 | 4 |
For details and further investigation, click here