| Full name: casein kinase 2 beta | Alias Symbol: Ckb1|Ckb2 | ||
| Type: protein-coding gene | Cytoband: 6p21.33 | ||
| Entrez ID: 1460 | HGNC ID: HGNC:2460 | Ensembl Gene: ENSG00000204435 | OMIM ID: 115441 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
CSNK2B involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04064 | NF-kappa B signaling pathway | |
| hsa04310 | Wnt signaling pathway | |
| hsa04520 | Adherens junction | |
| hsa04530 | Tight junction | |
| hsa05168 | Herpes simplex infection |
Expression of CSNK2B:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CSNK2B | 1460 | 201390_s_at | 0.4502 | 0.2533 | |
| GSE20347 | CSNK2B | 1460 | 201390_s_at | -0.3691 | 0.0093 | |
| GSE23400 | CSNK2B | 1460 | 201390_s_at | 0.0684 | 0.3151 | |
| GSE26886 | CSNK2B | 1460 | 201390_s_at | -0.8937 | 0.0001 | |
| GSE29001 | CSNK2B | 1460 | 201390_s_at | -0.3381 | 0.3266 | |
| GSE38129 | CSNK2B | 1460 | 201390_s_at | -0.1200 | 0.3366 | |
| GSE45670 | CSNK2B | 1460 | 201390_s_at | -0.1307 | 0.3535 | |
| GSE53622 | CSNK2B | 1460 | 61719 | -0.0877 | 0.1419 | |
| GSE53624 | CSNK2B | 1460 | 61719 | 0.1792 | 0.0002 | |
| GSE63941 | CSNK2B | 1460 | 201390_s_at | 1.3598 | 0.0003 | |
| GSE77861 | CSNK2B | 1460 | 201390_s_at | -0.4250 | 0.1325 | |
| GSE97050 | CSNK2B | 1460 | A_33_P3318027 | 0.2435 | 0.3820 | |
| SRP007169 | CSNK2B | 1460 | RNAseq | -0.9468 | 0.0166 | |
| SRP008496 | CSNK2B | 1460 | RNAseq | -0.8150 | 0.0001 | |
| SRP064894 | CSNK2B | 1460 | RNAseq | 0.2485 | 0.2930 | |
| SRP133303 | CSNK2B | 1460 | RNAseq | -0.0859 | 0.4829 | |
| SRP159526 | CSNK2B | 1460 | RNAseq | -0.0640 | 0.7219 | |
| SRP193095 | CSNK2B | 1460 | RNAseq | -0.1238 | 0.2390 | |
| SRP219564 | CSNK2B | 1460 | RNAseq | 0.1872 | 0.5189 | |
| TCGA | CSNK2B | 1460 | RNAseq | 0.1013 | 0.0255 |
Upregulated datasets: 1; Downregulated datasets: 0.
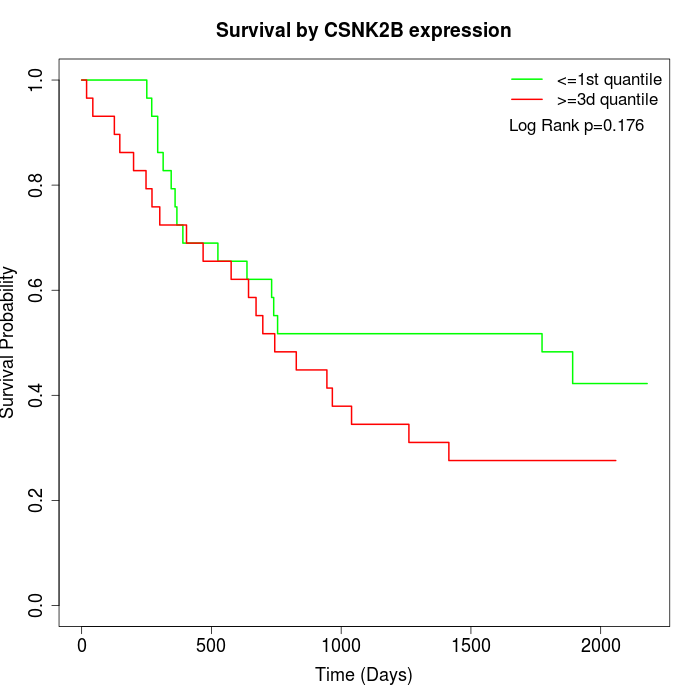
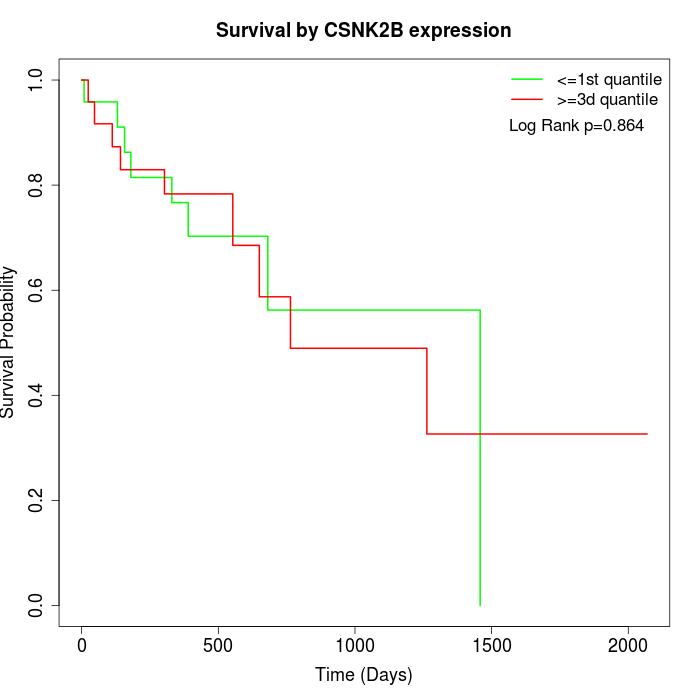
Survival by CSNK2B expression:
Note: Click image to view full size file.
Copy number change of CSNK2B:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CSNK2B | 1460 | 5 | 1 | 24 | |
| GSE20123 | CSNK2B | 1460 | 5 | 1 | 24 | |
| GSE43470 | CSNK2B | 1460 | 5 | 1 | 37 | |
| GSE46452 | CSNK2B | 1460 | 1 | 10 | 48 | |
| GSE47630 | CSNK2B | 1460 | 7 | 4 | 29 | |
| GSE54993 | CSNK2B | 1460 | 2 | 1 | 67 | |
| GSE54994 | CSNK2B | 1460 | 11 | 4 | 38 | |
| GSE60625 | CSNK2B | 1460 | 0 | 1 | 10 | |
| GSE74703 | CSNK2B | 1460 | 5 | 0 | 31 | |
| GSE74704 | CSNK2B | 1460 | 2 | 0 | 18 | |
| TCGA | CSNK2B | 1460 | 16 | 16 | 64 |
Total number of gains: 59; Total number of losses: 39; Total Number of normals: 390.
Somatic mutations of CSNK2B:
Generating mutation plots.
Highly correlated genes for CSNK2B:
Showing top 20/671 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CSNK2B | ERI3 | 0.836369 | 3 | 0 | 3 |
| CSNK2B | MRPL38 | 0.831402 | 3 | 0 | 3 |
| CSNK2B | SMARCC1 | 0.82144 | 3 | 0 | 3 |
| CSNK2B | ZKSCAN4 | 0.8203 | 3 | 0 | 3 |
| CSNK2B | LEMD2 | 0.813265 | 4 | 0 | 4 |
| CSNK2B | GPR160 | 0.799395 | 3 | 0 | 3 |
| CSNK2B | RPP38 | 0.788258 | 3 | 0 | 3 |
| CSNK2B | ERCC8 | 0.774695 | 3 | 0 | 3 |
| CSNK2B | KDM4A | 0.769094 | 3 | 0 | 3 |
| CSNK2B | COMMD2 | 0.74048 | 3 | 0 | 3 |
| CSNK2B | SFXN2 | 0.738419 | 3 | 0 | 3 |
| CSNK2B | CXCL16 | 0.737135 | 3 | 0 | 3 |
| CSNK2B | C12orf45 | 0.736361 | 3 | 0 | 3 |
| CSNK2B | RECQL5 | 0.734414 | 4 | 0 | 4 |
| CSNK2B | POLR2C | 0.732655 | 4 | 0 | 4 |
| CSNK2B | KDF1 | 0.730172 | 3 | 0 | 3 |
| CSNK2B | TMEM167A | 0.726237 | 3 | 0 | 3 |
| CSNK2B | HAVCR2 | 0.724219 | 3 | 0 | 3 |
| CSNK2B | MGAT4A | 0.715463 | 3 | 0 | 3 |
| CSNK2B | PHKB | 0.709075 | 3 | 0 | 3 |
For details and further investigation, click here