| Full name: C-X-C motif chemokine ligand 2 | Alias Symbol: SCYB2|GROb|MIP-2a|MGSA-b|CINC-2a | ||
| Type: protein-coding gene | Cytoband: 4q13.3 | ||
| Entrez ID: 2920 | HGNC ID: HGNC:4603 | Ensembl Gene: ENSG00000081041 | OMIM ID: 139110 |
| Drug and gene relationship at DGIdb | |||
CXCL2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04062 | Chemokine signaling pathway | |
| hsa04064 | NF-kappa B signaling pathway | |
| hsa05134 | Legionellosis |
Expression of CXCL2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CXCL2 | 2920 | 209774_x_at | 0.2404 | 0.9297 | |
| GSE20347 | CXCL2 | 2920 | 209774_x_at | 0.2197 | 0.6293 | |
| GSE23400 | CXCL2 | 2920 | 209774_x_at | -0.0569 | 0.7707 | |
| GSE26886 | CXCL2 | 2920 | 209774_x_at | 0.8800 | 0.3484 | |
| GSE29001 | CXCL2 | 2920 | 209774_x_at | 0.1871 | 0.7330 | |
| GSE38129 | CXCL2 | 2920 | 209774_x_at | -0.2076 | 0.6666 | |
| GSE45670 | CXCL2 | 2920 | 209774_x_at | 0.0717 | 0.8992 | |
| GSE53622 | CXCL2 | 2920 | 50463 | 1.2858 | 0.0000 | |
| GSE53624 | CXCL2 | 2920 | 50463 | 0.6896 | 0.0000 | |
| GSE63941 | CXCL2 | 2920 | 209774_x_at | -0.6400 | 0.7541 | |
| GSE77861 | CXCL2 | 2920 | 209774_x_at | 0.4671 | 0.2178 | |
| GSE97050 | CXCL2 | 2920 | A_24_P257416 | 1.2647 | 0.1372 | |
| SRP007169 | CXCL2 | 2920 | RNAseq | 0.0259 | 0.9798 | |
| SRP064894 | CXCL2 | 2920 | RNAseq | 1.0995 | 0.0049 | |
| SRP133303 | CXCL2 | 2920 | RNAseq | 0.9391 | 0.0204 | |
| SRP159526 | CXCL2 | 2920 | RNAseq | 1.1200 | 0.0013 | |
| SRP193095 | CXCL2 | 2920 | RNAseq | 1.2591 | 0.0021 | |
| SRP219564 | CXCL2 | 2920 | RNAseq | 1.6571 | 0.0335 | |
| TCGA | CXCL2 | 2920 | RNAseq | -0.2296 | 0.3004 |
Upregulated datasets: 5; Downregulated datasets: 0.
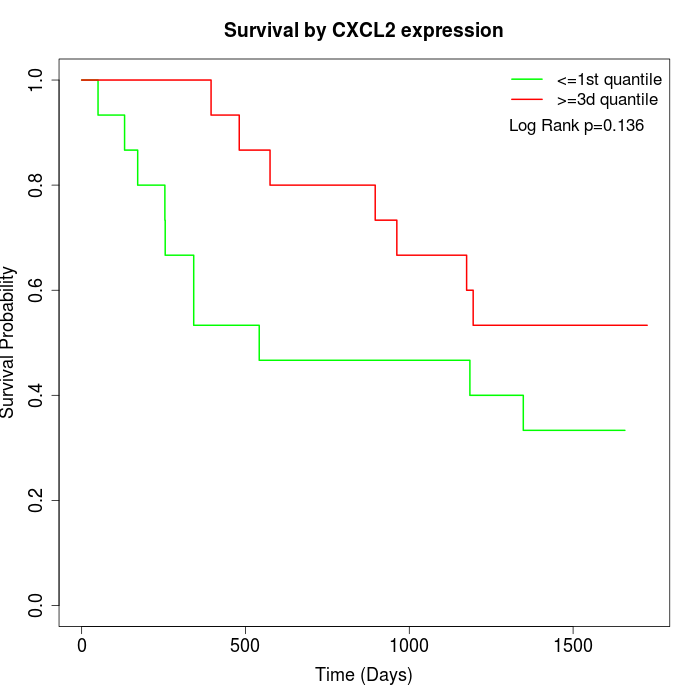
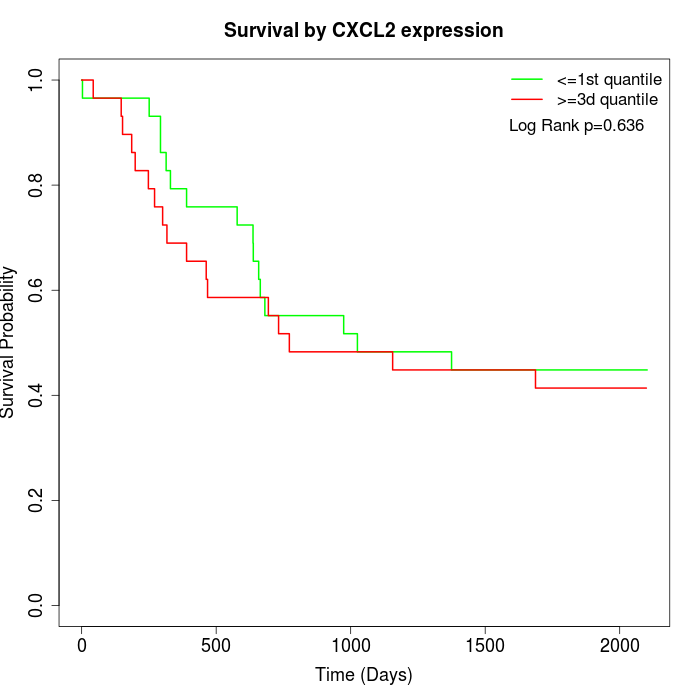
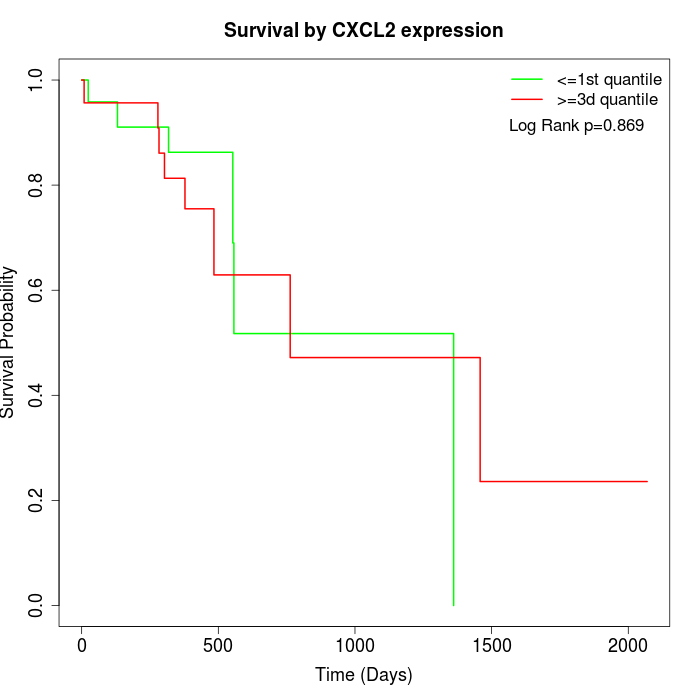
Survival by CXCL2 expression:
Note: Click image to view full size file.
Copy number change of CXCL2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CXCL2 | 2920 | 2 | 9 | 19 | |
| GSE20123 | CXCL2 | 2920 | 2 | 9 | 19 | |
| GSE43470 | CXCL2 | 2920 | 0 | 13 | 30 | |
| GSE46452 | CXCL2 | 2920 | 3 | 34 | 22 | |
| GSE47630 | CXCL2 | 2920 | 1 | 19 | 20 | |
| GSE54993 | CXCL2 | 2920 | 7 | 0 | 63 | |
| GSE54994 | CXCL2 | 2920 | 1 | 9 | 43 | |
| GSE60625 | CXCL2 | 2920 | 0 | 0 | 11 | |
| GSE74703 | CXCL2 | 2920 | 0 | 11 | 25 | |
| GSE74704 | CXCL2 | 2920 | 2 | 5 | 13 | |
| TCGA | CXCL2 | 2920 | 19 | 28 | 49 |
Total number of gains: 37; Total number of losses: 137; Total Number of normals: 314.
Somatic mutations of CXCL2:
Generating mutation plots.
Highly correlated genes for CXCL2:
Showing top 20/219 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CXCL2 | CXCL3 | 0.78658 | 12 | 0 | 12 |
| CXCL2 | ARL5B | 0.733549 | 3 | 0 | 3 |
| CXCL2 | EIF2AK2 | 0.72311 | 3 | 0 | 3 |
| CXCL2 | C9orf64 | 0.706585 | 3 | 0 | 3 |
| CXCL2 | ZNF275 | 0.704054 | 3 | 0 | 3 |
| CXCL2 | RPGR | 0.70227 | 3 | 0 | 3 |
| CXCL2 | SLC25A16 | 0.668737 | 3 | 0 | 3 |
| CXCL2 | CXCL1 | 0.666827 | 11 | 0 | 8 |
| CXCL2 | SLC26A6 | 0.66557 | 3 | 0 | 3 |
| CXCL2 | GSTO1 | 0.663309 | 3 | 0 | 3 |
| CXCL2 | MZT2B | 0.654144 | 3 | 0 | 3 |
| CXCL2 | TMX3 | 0.638998 | 3 | 0 | 3 |
| CXCL2 | APOC1 | 0.638748 | 4 | 0 | 4 |
| CXCL2 | SH2B2 | 0.638613 | 3 | 0 | 3 |
| CXCL2 | DLGAP4 | 0.637479 | 4 | 0 | 3 |
| CXCL2 | ICAM1 | 0.63657 | 8 | 0 | 6 |
| CXCL2 | TNFAIP3 | 0.632934 | 13 | 0 | 11 |
| CXCL2 | CXCL8 | 0.628697 | 10 | 0 | 6 |
| CXCL2 | ZC3H12A | 0.628259 | 4 | 0 | 3 |
| CXCL2 | HSP90B1 | 0.627444 | 4 | 0 | 4 |
For details and further investigation, click here