| Full name: C-X-C motif chemokine ligand 1 | Alias Symbol: SCYB1|GROa|MGSA-a|NAP-3 | ||
| Type: protein-coding gene | Cytoband: 4q13.3 | ||
| Entrez ID: 2919 | HGNC ID: HGNC:4602 | Ensembl Gene: ENSG00000163739 | OMIM ID: 155730 |
| Drug and gene relationship at DGIdb | |||
CXCL1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04062 | Chemokine signaling pathway | |
| hsa05120 | Epithelial cell signaling in Helicobacter pylori infection | |
| hsa05134 | Legionellosis |
Expression of CXCL1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CXCL1 | 2919 | 204470_at | 2.4371 | 0.2189 | |
| GSE20347 | CXCL1 | 2919 | 204470_at | 1.7546 | 0.0007 | |
| GSE23400 | CXCL1 | 2919 | 204470_at | 1.2652 | 0.0000 | |
| GSE26886 | CXCL1 | 2919 | 204470_at | 2.3541 | 0.0168 | |
| GSE29001 | CXCL1 | 2919 | 204470_at | 2.1141 | 0.0002 | |
| GSE38129 | CXCL1 | 2919 | 204470_at | 2.0608 | 0.0000 | |
| GSE45670 | CXCL1 | 2919 | 204470_at | 3.9362 | 0.0000 | |
| GSE53622 | CXCL1 | 2919 | 144640 | 2.3168 | 0.0000 | |
| GSE53624 | CXCL1 | 2919 | 144640 | 1.8558 | 0.0000 | |
| GSE63941 | CXCL1 | 2919 | 204470_at | -3.1538 | 0.0656 | |
| GSE77861 | CXCL1 | 2919 | 204470_at | 2.2891 | 0.0036 | |
| GSE97050 | CXCL1 | 2919 | A_33_P3330264 | 1.8941 | 0.1241 | |
| SRP007169 | CXCL1 | 2919 | RNAseq | 3.4056 | 0.0002 | |
| SRP064894 | CXCL1 | 2919 | RNAseq | 3.4464 | 0.0000 | |
| SRP133303 | CXCL1 | 2919 | RNAseq | 2.2589 | 0.0001 | |
| SRP159526 | CXCL1 | 2919 | RNAseq | 4.3525 | 0.0000 | |
| SRP193095 | CXCL1 | 2919 | RNAseq | 3.5246 | 0.0000 | |
| SRP219564 | CXCL1 | 2919 | RNAseq | 3.1625 | 0.0005 | |
| TCGA | CXCL1 | 2919 | RNAseq | 1.5411 | 0.0000 |
Upregulated datasets: 16; Downregulated datasets: 0.
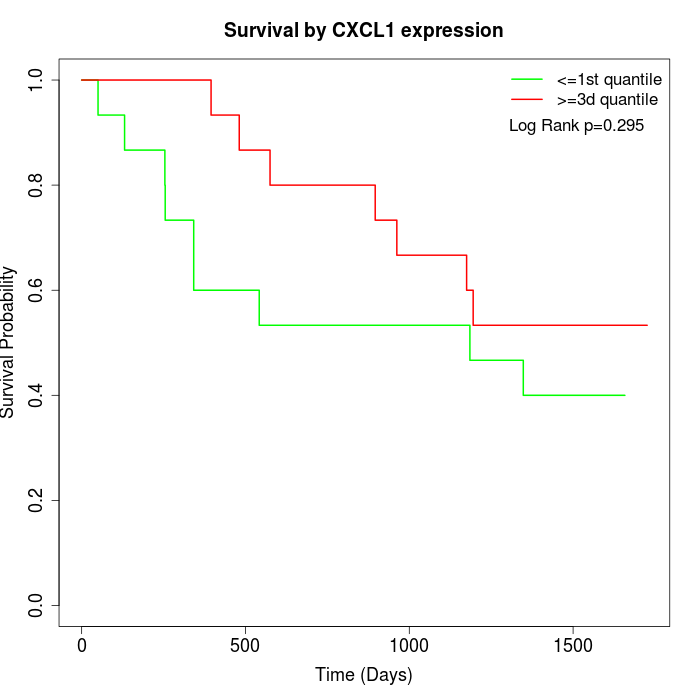
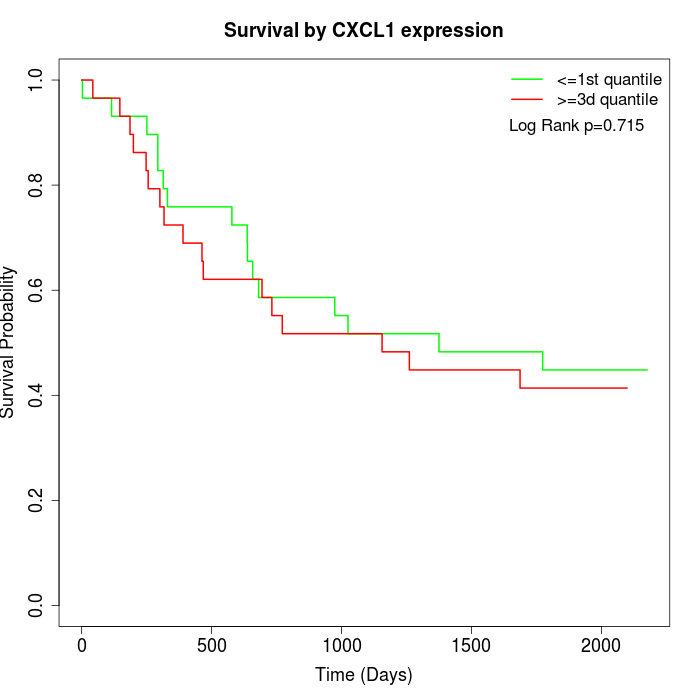
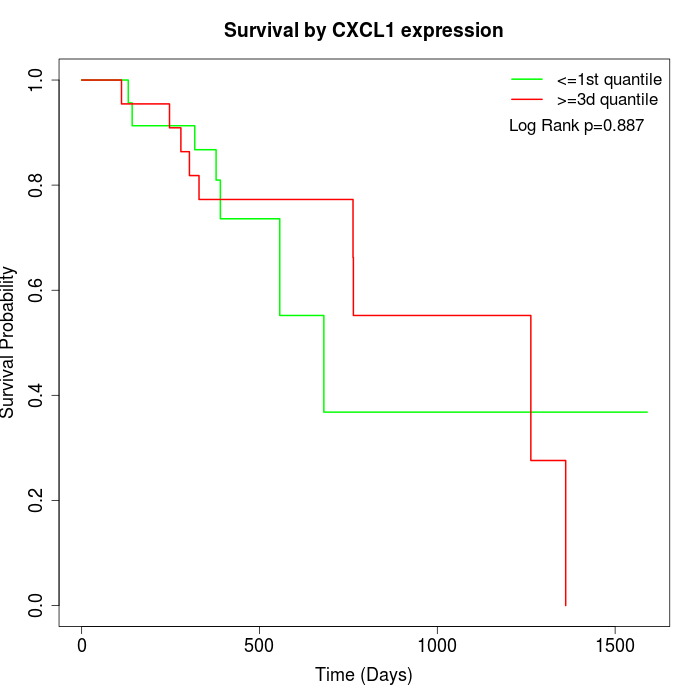
Survival by CXCL1 expression:
Note: Click image to view full size file.
Copy number change of CXCL1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CXCL1 | 2919 | 2 | 9 | 19 | |
| GSE20123 | CXCL1 | 2919 | 2 | 9 | 19 | |
| GSE43470 | CXCL1 | 2919 | 0 | 13 | 30 | |
| GSE46452 | CXCL1 | 2919 | 3 | 34 | 22 | |
| GSE47630 | CXCL1 | 2919 | 1 | 19 | 20 | |
| GSE54993 | CXCL1 | 2919 | 7 | 0 | 63 | |
| GSE54994 | CXCL1 | 2919 | 1 | 9 | 43 | |
| GSE60625 | CXCL1 | 2919 | 0 | 0 | 11 | |
| GSE74703 | CXCL1 | 2919 | 0 | 11 | 25 | |
| GSE74704 | CXCL1 | 2919 | 2 | 5 | 13 | |
| TCGA | CXCL1 | 2919 | 19 | 28 | 49 |
Total number of gains: 37; Total number of losses: 137; Total Number of normals: 314.
Somatic mutations of CXCL1:
Generating mutation plots.
Highly correlated genes for CXCL1:
Showing top 20/1148 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CXCL1 | CXCL8 | 0.850738 | 11 | 0 | 11 |
| CXCL1 | CXCL6 | 0.817794 | 12 | 0 | 12 |
| CXCL1 | LRRC8C | 0.744854 | 3 | 0 | 3 |
| CXCL1 | MMP12 | 0.744075 | 11 | 0 | 11 |
| CXCL1 | IKBIP | 0.737732 | 6 | 0 | 6 |
| CXCL1 | UHMK1 | 0.735743 | 4 | 0 | 4 |
| CXCL1 | ABCA13 | 0.725931 | 3 | 0 | 3 |
| CXCL1 | CDCA2 | 0.724701 | 5 | 0 | 5 |
| CXCL1 | FAM89A | 0.713442 | 3 | 0 | 3 |
| CXCL1 | TMEM138 | 0.711208 | 6 | 0 | 6 |
| CXCL1 | MET | 0.700338 | 10 | 0 | 9 |
| CXCL1 | XPR1 | 0.699838 | 7 | 0 | 6 |
| CXCL1 | RUNX2 | 0.69854 | 5 | 0 | 5 |
| CXCL1 | SPRY4-IT1 | 0.698504 | 3 | 0 | 3 |
| CXCL1 | TNFRSF10B | 0.696613 | 11 | 0 | 11 |
| CXCL1 | C11orf24 | 0.695659 | 10 | 0 | 9 |
| CXCL1 | FCGR3A | 0.687684 | 3 | 0 | 3 |
| CXCL1 | MTHFD2 | 0.684298 | 11 | 0 | 11 |
| CXCL1 | PDIA5 | 0.682154 | 3 | 0 | 3 |
| CXCL1 | CLSPN | 0.680024 | 7 | 0 | 7 |
For details and further investigation, click here