| Full name: cytochrome P450 family 27 subfamily A member 1 | Alias Symbol: CTX|CP27 | ||
| Type: protein-coding gene | Cytoband: 2q35 | ||
| Entrez ID: 1593 | HGNC ID: HGNC:2605 | Ensembl Gene: ENSG00000135929 | OMIM ID: 606530 |
| Drug and gene relationship at DGIdb | |||
CYP27A1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa03320 | PPAR signaling pathway |
Expression of CYP27A1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CYP27A1 | 1593 | 203979_at | -0.2323 | 0.5604 | |
| GSE20347 | CYP27A1 | 1593 | 203979_at | -0.0733 | 0.5479 | |
| GSE23400 | CYP27A1 | 1593 | 203979_at | -0.0704 | 0.2544 | |
| GSE26886 | CYP27A1 | 1593 | 203979_at | 0.1694 | 0.2168 | |
| GSE29001 | CYP27A1 | 1593 | 203979_at | -0.2138 | 0.2463 | |
| GSE38129 | CYP27A1 | 1593 | 203979_at | -0.1921 | 0.0786 | |
| GSE45670 | CYP27A1 | 1593 | 203979_at | -0.3973 | 0.0000 | |
| GSE53622 | CYP27A1 | 1593 | 9047 | -0.3843 | 0.0252 | |
| GSE53624 | CYP27A1 | 1593 | 9047 | -0.5232 | 0.0010 | |
| GSE63941 | CYP27A1 | 1593 | 203979_at | -0.9147 | 0.0001 | |
| GSE77861 | CYP27A1 | 1593 | 203979_at | -0.1799 | 0.1711 | |
| GSE97050 | CYP27A1 | 1593 | A_33_P3361422 | -0.3222 | 0.3486 | |
| SRP007169 | CYP27A1 | 1593 | RNAseq | 3.4989 | 0.0028 | |
| SRP064894 | CYP27A1 | 1593 | RNAseq | 0.4040 | 0.3825 | |
| SRP133303 | CYP27A1 | 1593 | RNAseq | -0.9202 | 0.0031 | |
| SRP159526 | CYP27A1 | 1593 | RNAseq | 1.2792 | 0.0733 | |
| SRP193095 | CYP27A1 | 1593 | RNAseq | -0.9679 | 0.0092 | |
| SRP219564 | CYP27A1 | 1593 | RNAseq | -0.1482 | 0.8486 | |
| TCGA | CYP27A1 | 1593 | RNAseq | -0.5428 | 0.0037 |
Upregulated datasets: 1; Downregulated datasets: 0.
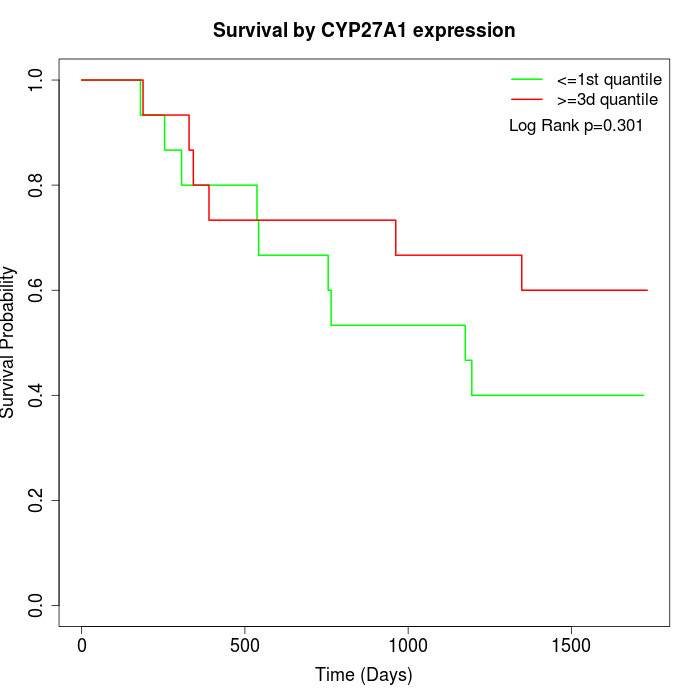
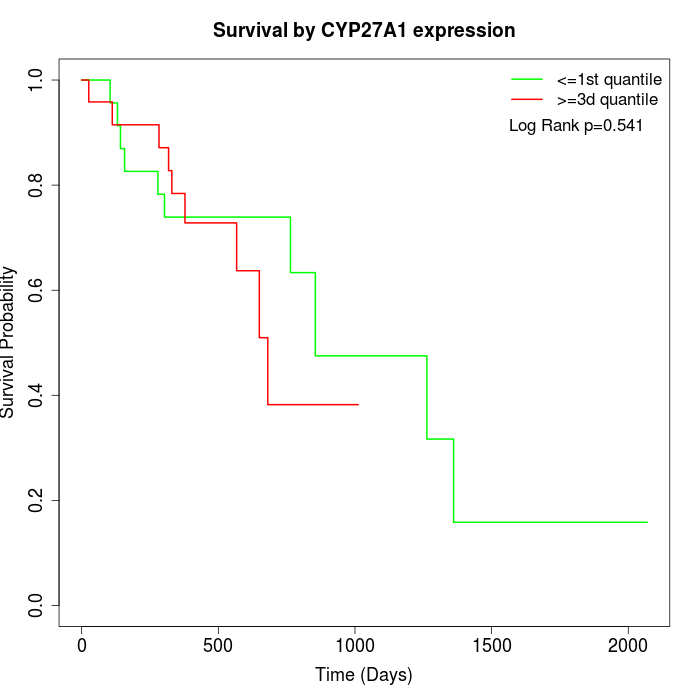
Survival by CYP27A1 expression:
Note: Click image to view full size file.
Copy number change of CYP27A1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CYP27A1 | 1593 | 1 | 14 | 15 | |
| GSE20123 | CYP27A1 | 1593 | 1 | 13 | 16 | |
| GSE43470 | CYP27A1 | 1593 | 1 | 7 | 35 | |
| GSE46452 | CYP27A1 | 1593 | 0 | 5 | 54 | |
| GSE47630 | CYP27A1 | 1593 | 4 | 5 | 31 | |
| GSE54993 | CYP27A1 | 1593 | 1 | 2 | 67 | |
| GSE54994 | CYP27A1 | 1593 | 7 | 10 | 36 | |
| GSE60625 | CYP27A1 | 1593 | 0 | 3 | 8 | |
| GSE74703 | CYP27A1 | 1593 | 1 | 5 | 30 | |
| GSE74704 | CYP27A1 | 1593 | 1 | 7 | 12 | |
| TCGA | CYP27A1 | 1593 | 11 | 27 | 58 |
Total number of gains: 28; Total number of losses: 98; Total Number of normals: 362.
Somatic mutations of CYP27A1:
Generating mutation plots.
Highly correlated genes for CYP27A1:
Showing top 20/774 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CYP27A1 | AFF3 | 0.794856 | 3 | 0 | 3 |
| CYP27A1 | MAPRE3 | 0.79463 | 3 | 0 | 3 |
| CYP27A1 | MED25 | 0.773357 | 3 | 0 | 3 |
| CYP27A1 | C20orf194 | 0.764066 | 3 | 0 | 3 |
| CYP27A1 | LRRC34 | 0.750819 | 3 | 0 | 3 |
| CYP27A1 | FOXO3 | 0.743278 | 4 | 0 | 4 |
| CYP27A1 | GBP2 | 0.74035 | 3 | 0 | 3 |
| CYP27A1 | RPL32 | 0.737736 | 4 | 0 | 4 |
| CYP27A1 | KPNA6 | 0.734956 | 3 | 0 | 3 |
| CYP27A1 | ST3GAL2 | 0.733727 | 3 | 0 | 3 |
| CYP27A1 | SETD7 | 0.727942 | 3 | 0 | 3 |
| CYP27A1 | ARHGAP31 | 0.726225 | 5 | 0 | 5 |
| CYP27A1 | MICA | 0.725133 | 3 | 0 | 3 |
| CYP27A1 | PINK1 | 0.72194 | 3 | 0 | 3 |
| CYP27A1 | FIBIN | 0.720903 | 5 | 0 | 5 |
| CYP27A1 | UBE4A | 0.717527 | 3 | 0 | 3 |
| CYP27A1 | ZNF23 | 0.714451 | 3 | 0 | 3 |
| CYP27A1 | SNRK | 0.7098 | 3 | 0 | 3 |
| CYP27A1 | EIF1AY | 0.707265 | 5 | 0 | 4 |
| CYP27A1 | CLDN2 | 0.704685 | 3 | 0 | 3 |
For details and further investigation, click here