| Full name: protein kinase C eta | Alias Symbol: PKC-L|PKCL | ||
| Type: protein-coding gene | Cytoband: 14q23.1 | ||
| Entrez ID: 5583 | HGNC ID: HGNC:9403 | Ensembl Gene: ENSG00000027075 | OMIM ID: 605437 |
| Drug and gene relationship at DGIdb | |||
PRKCH involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04270 | Vascular smooth muscle contraction | |
| hsa04530 | Tight junction | |
| hsa04750 | Inflammatory mediator regulation of TRP channels |
Expression of PRKCH:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PRKCH | 5583 | 218764_at | 0.1284 | 0.8979 | |
| GSE20347 | PRKCH | 5583 | 218764_at | -0.8090 | 0.0001 | |
| GSE23400 | PRKCH | 5583 | 218764_at | -0.6115 | 0.0000 | |
| GSE26886 | PRKCH | 5583 | 218764_at | -1.4846 | 0.0000 | |
| GSE29001 | PRKCH | 5583 | 218764_at | -1.1004 | 0.0112 | |
| GSE38129 | PRKCH | 5583 | 218764_at | -0.6487 | 0.0010 | |
| GSE45670 | PRKCH | 5583 | 218764_at | -0.3973 | 0.0495 | |
| GSE53622 | PRKCH | 5583 | 129297 | -0.6556 | 0.0000 | |
| GSE53624 | PRKCH | 5583 | 129297 | -0.9137 | 0.0000 | |
| GSE63941 | PRKCH | 5583 | 218764_at | 2.7146 | 0.0049 | |
| GSE77861 | PRKCH | 5583 | 218764_at | -0.8507 | 0.0054 | |
| GSE97050 | PRKCH | 5583 | A_23_P205567 | 0.2288 | 0.4572 | |
| SRP007169 | PRKCH | 5583 | RNAseq | -1.3717 | 0.0002 | |
| SRP008496 | PRKCH | 5583 | RNAseq | -1.4040 | 0.0000 | |
| SRP064894 | PRKCH | 5583 | RNAseq | -0.8240 | 0.0004 | |
| SRP133303 | PRKCH | 5583 | RNAseq | -0.8106 | 0.0000 | |
| SRP159526 | PRKCH | 5583 | RNAseq | -1.0101 | 0.0713 | |
| SRP193095 | PRKCH | 5583 | RNAseq | -0.7974 | 0.0000 | |
| SRP219564 | PRKCH | 5583 | RNAseq | -0.7460 | 0.1482 | |
| TCGA | PRKCH | 5583 | RNAseq | 0.1534 | 0.0149 |
Upregulated datasets: 1; Downregulated datasets: 4.
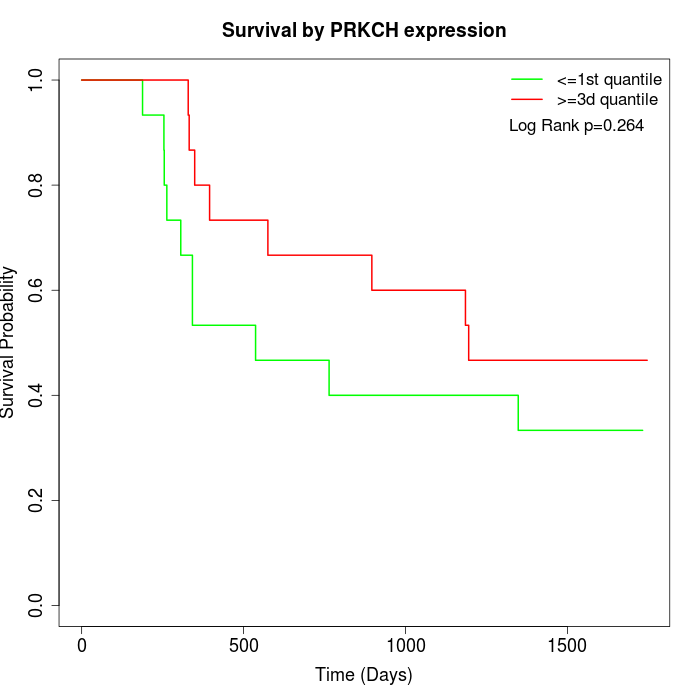
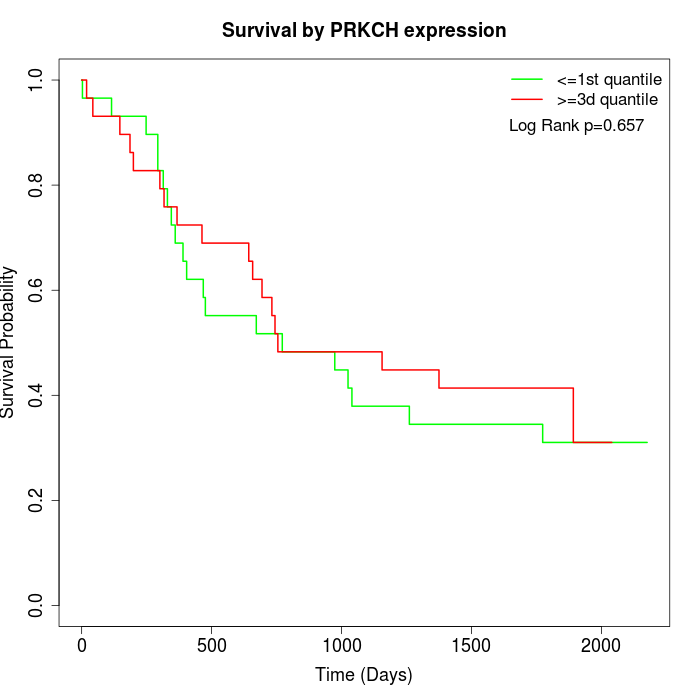
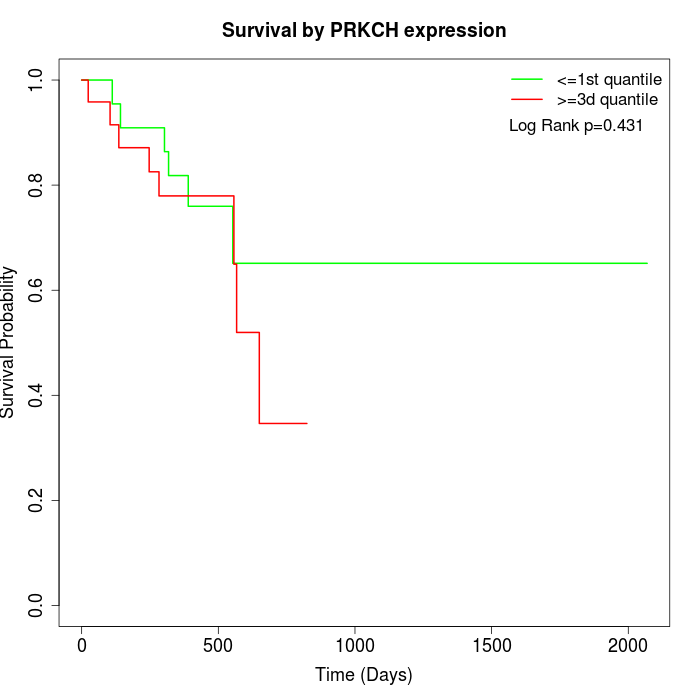
Survival by PRKCH expression:
Note: Click image to view full size file.
Copy number change of PRKCH:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PRKCH | 5583 | 9 | 4 | 17 | |
| GSE20123 | PRKCH | 5583 | 9 | 4 | 17 | |
| GSE43470 | PRKCH | 5583 | 8 | 2 | 33 | |
| GSE46452 | PRKCH | 5583 | 16 | 3 | 40 | |
| GSE47630 | PRKCH | 5583 | 11 | 9 | 20 | |
| GSE54993 | PRKCH | 5583 | 3 | 9 | 58 | |
| GSE54994 | PRKCH | 5583 | 19 | 4 | 30 | |
| GSE60625 | PRKCH | 5583 | 0 | 2 | 9 | |
| GSE74703 | PRKCH | 5583 | 7 | 2 | 27 | |
| GSE74704 | PRKCH | 5583 | 4 | 3 | 13 | |
| TCGA | PRKCH | 5583 | 32 | 15 | 49 |
Total number of gains: 118; Total number of losses: 57; Total Number of normals: 313.
Somatic mutations of PRKCH:
Generating mutation plots.
Highly correlated genes for PRKCH:
Showing top 20/1375 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PRKCH | IER3IP1 | 0.8057 | 3 | 0 | 3 |
| PRKCH | ARPC1A | 0.801155 | 4 | 0 | 4 |
| PRKCH | PGM2 | 0.783888 | 5 | 0 | 5 |
| PRKCH | DNAJB7 | 0.778578 | 3 | 0 | 3 |
| PRKCH | ERCC6 | 0.775217 | 4 | 0 | 4 |
| PRKCH | CNOT11 | 0.774326 | 3 | 0 | 3 |
| PRKCH | BROX | 0.773665 | 3 | 0 | 3 |
| PRKCH | DDI2 | 0.764029 | 3 | 0 | 3 |
| PRKCH | MAL2 | 0.755966 | 8 | 0 | 7 |
| PRKCH | GPR108 | 0.740503 | 3 | 0 | 3 |
| PRKCH | TOM1 | 0.740337 | 10 | 0 | 9 |
| PRKCH | RASSF5 | 0.739328 | 7 | 0 | 6 |
| PRKCH | SAMD12 | 0.737124 | 3 | 0 | 3 |
| PRKCH | ASCC2 | 0.733501 | 11 | 0 | 11 |
| PRKCH | CDKN2C | 0.733229 | 3 | 0 | 3 |
| PRKCH | ENOPH1 | 0.731487 | 3 | 0 | 3 |
| PRKCH | OSTF1 | 0.731352 | 12 | 0 | 11 |
| PRKCH | VSIG10L | 0.731314 | 5 | 0 | 5 |
| PRKCH | LRP10 | 0.73079 | 11 | 0 | 10 |
| PRKCH | TPRG1 | 0.730082 | 8 | 0 | 8 |
For details and further investigation, click here