| Full name: dihydropyrimidinase like 2 | Alias Symbol: DRP-2|DHPRP2|CRMP2|DRP2 | ||
| Type: protein-coding gene | Cytoband: 8p21.2 | ||
| Entrez ID: 1808 | HGNC ID: HGNC:3014 | Ensembl Gene: ENSG00000092964 | OMIM ID: 602463 |
| Drug and gene relationship at DGIdb | |||
DPYSL2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04360 | Axon guidance |
Expression of DPYSL2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | DPYSL2 | 1808 | 200762_at | -0.5762 | 0.5013 | |
| GSE20347 | DPYSL2 | 1808 | 200762_at | 0.8925 | 0.0124 | |
| GSE23400 | DPYSL2 | 1808 | 200762_at | 0.1770 | 0.3714 | |
| GSE26886 | DPYSL2 | 1808 | 200762_at | 1.5997 | 0.0009 | |
| GSE29001 | DPYSL2 | 1808 | 200762_at | 1.0836 | 0.0078 | |
| GSE38129 | DPYSL2 | 1808 | 200762_at | 0.2387 | 0.5399 | |
| GSE45670 | DPYSL2 | 1808 | 200762_at | -1.1192 | 0.0020 | |
| GSE53622 | DPYSL2 | 1808 | 98632 | -0.2570 | 0.0555 | |
| GSE53624 | DPYSL2 | 1808 | 35245 | 0.1106 | 0.2465 | |
| GSE63941 | DPYSL2 | 1808 | 200762_at | -3.8038 | 0.0006 | |
| GSE77861 | DPYSL2 | 1808 | 200762_at | 1.7649 | 0.0012 | |
| GSE97050 | DPYSL2 | 1808 | A_24_P38347 | -0.0267 | 0.9410 | |
| SRP007169 | DPYSL2 | 1808 | RNAseq | 1.9047 | 0.0001 | |
| SRP008496 | DPYSL2 | 1808 | RNAseq | 1.8467 | 0.0000 | |
| SRP064894 | DPYSL2 | 1808 | RNAseq | 0.3581 | 0.2620 | |
| SRP133303 | DPYSL2 | 1808 | RNAseq | -0.3774 | 0.0893 | |
| SRP159526 | DPYSL2 | 1808 | RNAseq | 0.9103 | 0.0627 | |
| SRP193095 | DPYSL2 | 1808 | RNAseq | 1.2363 | 0.0000 | |
| SRP219564 | DPYSL2 | 1808 | RNAseq | 0.2783 | 0.6649 | |
| TCGA | DPYSL2 | 1808 | RNAseq | -0.1771 | 0.0344 |
Upregulated datasets: 6; Downregulated datasets: 2.
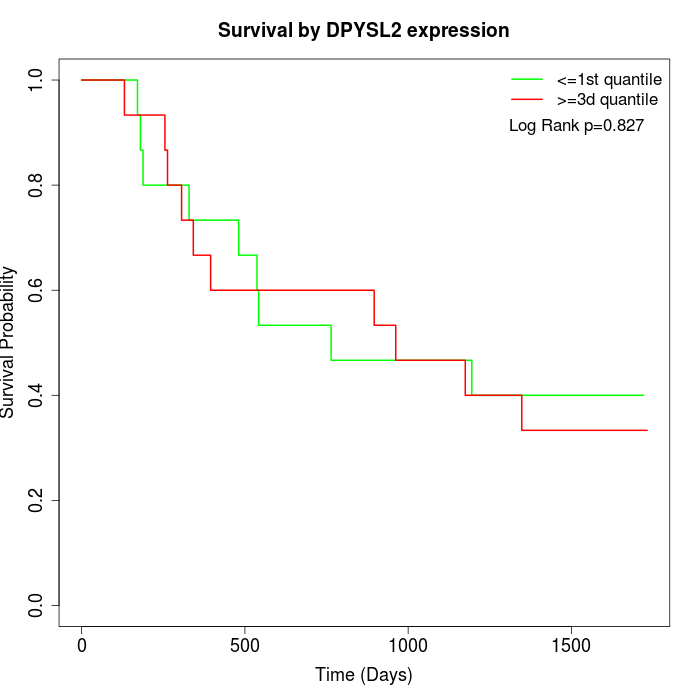
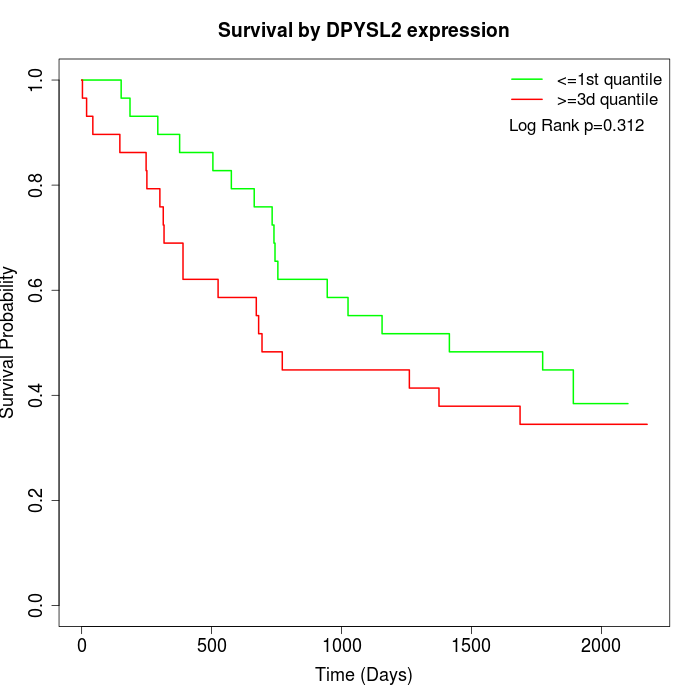
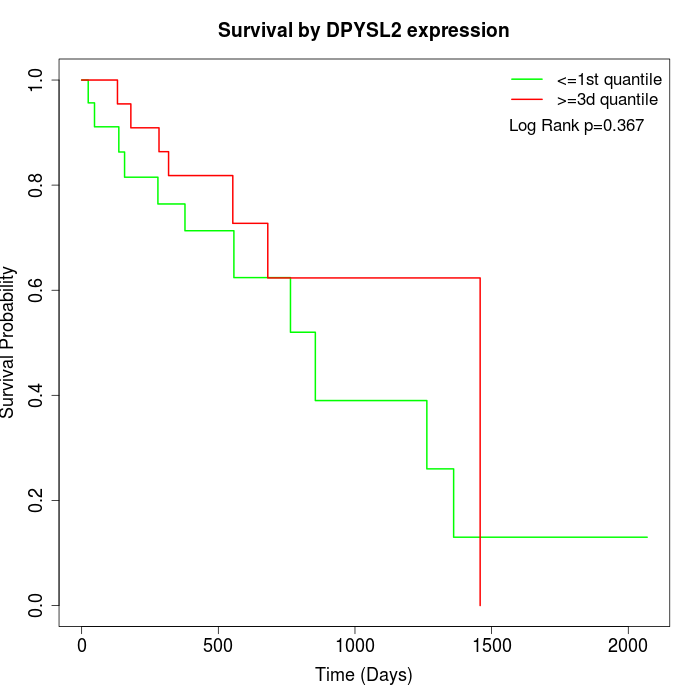
Survival by DPYSL2 expression:
Note: Click image to view full size file.
Copy number change of DPYSL2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | DPYSL2 | 1808 | 4 | 12 | 14 | |
| GSE20123 | DPYSL2 | 1808 | 4 | 12 | 14 | |
| GSE43470 | DPYSL2 | 1808 | 4 | 7 | 32 | |
| GSE46452 | DPYSL2 | 1808 | 13 | 13 | 33 | |
| GSE47630 | DPYSL2 | 1808 | 10 | 8 | 22 | |
| GSE54993 | DPYSL2 | 1808 | 2 | 15 | 53 | |
| GSE54994 | DPYSL2 | 1808 | 9 | 16 | 28 | |
| GSE60625 | DPYSL2 | 1808 | 3 | 0 | 8 | |
| GSE74703 | DPYSL2 | 1808 | 4 | 6 | 26 | |
| GSE74704 | DPYSL2 | 1808 | 3 | 8 | 9 | |
| TCGA | DPYSL2 | 1808 | 15 | 44 | 37 |
Total number of gains: 71; Total number of losses: 141; Total Number of normals: 276.
Somatic mutations of DPYSL2:
Generating mutation plots.
Highly correlated genes for DPYSL2:
Showing top 20/1303 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| DPYSL2 | ATIC | 0.819258 | 3 | 0 | 3 |
| DPYSL2 | GNPDA1 | 0.7944 | 3 | 0 | 3 |
| DPYSL2 | PAK1 | 0.792667 | 3 | 0 | 3 |
| DPYSL2 | PHLDB2 | 0.78691 | 3 | 0 | 3 |
| DPYSL2 | KIF13A | 0.783161 | 3 | 0 | 3 |
| DPYSL2 | TNFRSF1A | 0.78297 | 3 | 0 | 3 |
| DPYSL2 | PLBD2 | 0.782624 | 3 | 0 | 3 |
| DPYSL2 | LRRC59 | 0.769337 | 3 | 0 | 3 |
| DPYSL2 | FBN1 | 0.752608 | 11 | 0 | 10 |
| DPYSL2 | C11orf96 | 0.746378 | 5 | 0 | 5 |
| DPYSL2 | PIF1 | 0.744526 | 3 | 0 | 3 |
| DPYSL2 | TRRAP | 0.743854 | 4 | 0 | 4 |
| DPYSL2 | BICC1 | 0.743104 | 9 | 0 | 9 |
| DPYSL2 | LINC01279 | 0.741236 | 4 | 0 | 4 |
| DPYSL2 | ZC3H12C | 0.739391 | 3 | 0 | 3 |
| DPYSL2 | SGCE | 0.732752 | 11 | 0 | 10 |
| DPYSL2 | PMP22 | 0.726719 | 13 | 0 | 11 |
| DPYSL2 | TMEM38B | 0.725677 | 3 | 0 | 3 |
| DPYSL2 | ABCB10 | 0.721581 | 3 | 0 | 3 |
| DPYSL2 | DNM1L | 0.721394 | 3 | 0 | 3 |
For details and further investigation, click here