| Full name: dual specificity phosphatase 11 | Alias Symbol: PIR1 | ||
| Type: protein-coding gene | Cytoband: 2p13.1 | ||
| Entrez ID: 8446 | HGNC ID: HGNC:3066 | Ensembl Gene: ENSG00000144048 | OMIM ID: 603092 |
| Drug and gene relationship at DGIdb | |||
Expression of DUSP11:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | DUSP11 | 8446 | 202703_at | -0.1493 | 0.8163 | |
| GSE20347 | DUSP11 | 8446 | 202703_at | -0.2227 | 0.0348 | |
| GSE23400 | DUSP11 | 8446 | 202703_at | 0.0329 | 0.6761 | |
| GSE26886 | DUSP11 | 8446 | 202703_at | -0.9736 | 0.0000 | |
| GSE29001 | DUSP11 | 8446 | 202703_at | -0.2809 | 0.2812 | |
| GSE38129 | DUSP11 | 8446 | 202703_at | 0.1163 | 0.5287 | |
| GSE45670 | DUSP11 | 8446 | 202703_at | 0.0375 | 0.7672 | |
| GSE53622 | DUSP11 | 8446 | 38176 | 0.1958 | 0.0082 | |
| GSE53624 | DUSP11 | 8446 | 38176 | -0.0352 | 0.4558 | |
| GSE63941 | DUSP11 | 8446 | 202703_at | 0.3309 | 0.4403 | |
| GSE77861 | DUSP11 | 8446 | 202703_at | -0.3276 | 0.1928 | |
| GSE97050 | DUSP11 | 8446 | A_23_P376759 | 0.4808 | 0.1784 | |
| SRP007169 | DUSP11 | 8446 | RNAseq | -0.9714 | 0.0101 | |
| SRP008496 | DUSP11 | 8446 | RNAseq | -0.9317 | 0.0000 | |
| SRP064894 | DUSP11 | 8446 | RNAseq | -0.3555 | 0.0561 | |
| SRP133303 | DUSP11 | 8446 | RNAseq | 0.0493 | 0.7768 | |
| SRP159526 | DUSP11 | 8446 | RNAseq | -0.3342 | 0.1072 | |
| SRP193095 | DUSP11 | 8446 | RNAseq | -0.4701 | 0.0023 | |
| SRP219564 | DUSP11 | 8446 | RNAseq | -0.3247 | 0.5147 | |
| TCGA | DUSP11 | 8446 | RNAseq | 0.2839 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 0.
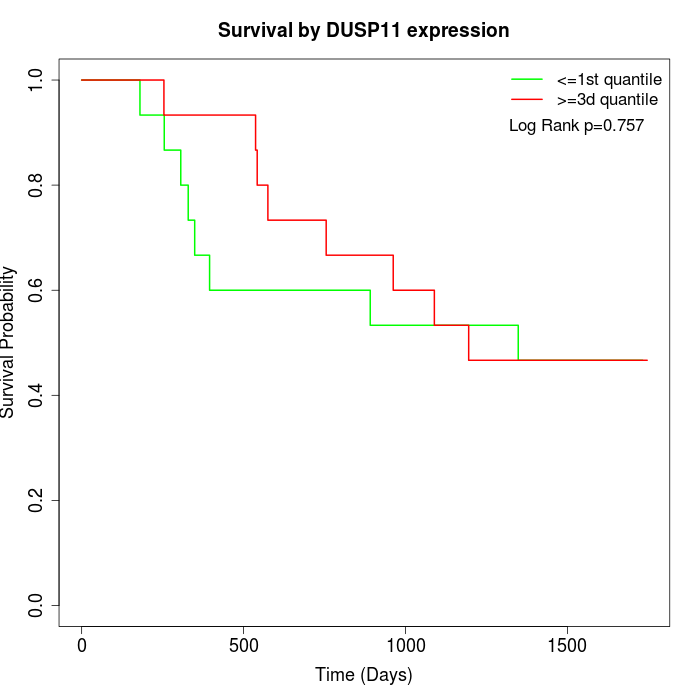
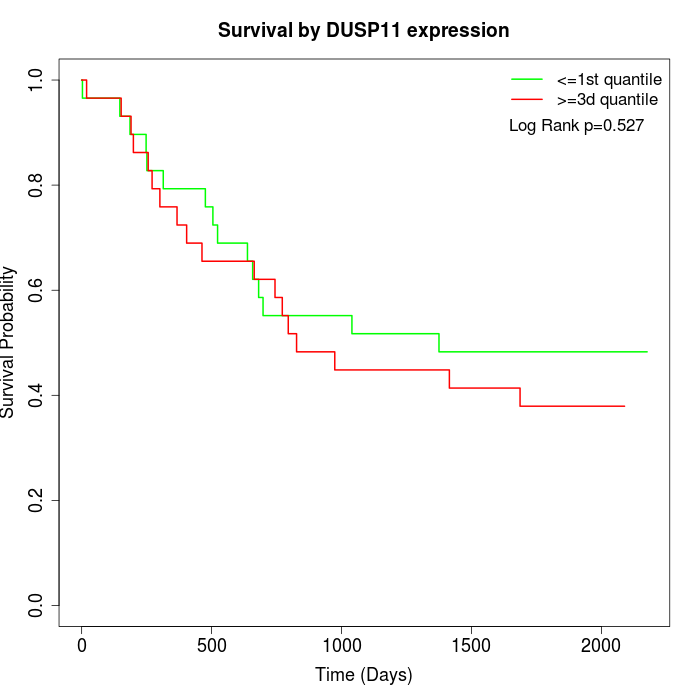
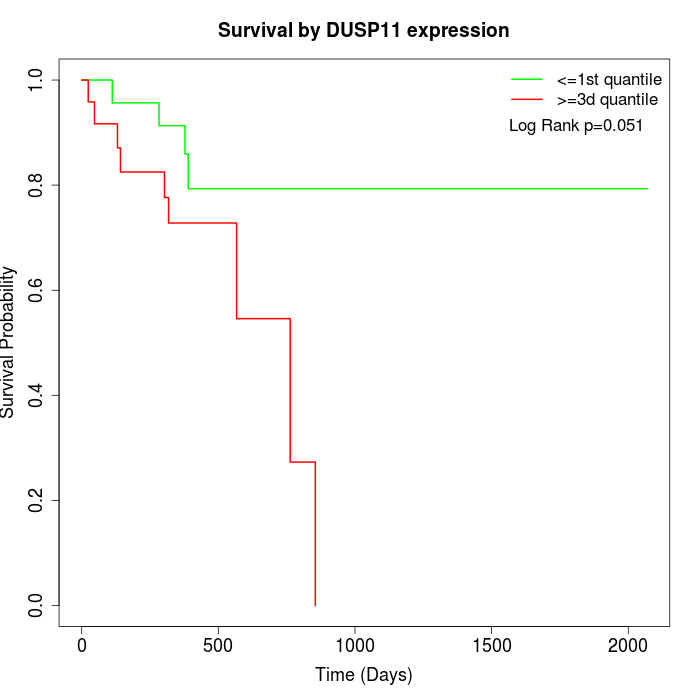
Survival by DUSP11 expression:
Note: Click image to view full size file.
Copy number change of DUSP11:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | DUSP11 | 8446 | 10 | 1 | 19 | |
| GSE20123 | DUSP11 | 8446 | 10 | 1 | 19 | |
| GSE43470 | DUSP11 | 8446 | 4 | 0 | 39 | |
| GSE46452 | DUSP11 | 8446 | 2 | 3 | 54 | |
| GSE47630 | DUSP11 | 8446 | 7 | 0 | 33 | |
| GSE54993 | DUSP11 | 8446 | 0 | 6 | 64 | |
| GSE54994 | DUSP11 | 8446 | 11 | 0 | 42 | |
| GSE60625 | DUSP11 | 8446 | 0 | 3 | 8 | |
| GSE74703 | DUSP11 | 8446 | 4 | 0 | 32 | |
| GSE74704 | DUSP11 | 8446 | 8 | 0 | 12 | |
| TCGA | DUSP11 | 8446 | 38 | 2 | 56 |
Total number of gains: 94; Total number of losses: 16; Total Number of normals: 378.
Somatic mutations of DUSP11:
Generating mutation plots.
Highly correlated genes for DUSP11:
Showing top 20/639 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| DUSP11 | GSKIP | 0.797639 | 3 | 0 | 3 |
| DUSP11 | PPP2CB | 0.731045 | 3 | 0 | 3 |
| DUSP11 | COBLL1 | 0.707783 | 3 | 0 | 3 |
| DUSP11 | CCNDBP1 | 0.706187 | 3 | 0 | 3 |
| DUSP11 | GIN1 | 0.702209 | 3 | 0 | 3 |
| DUSP11 | RTCA | 0.697924 | 7 | 0 | 7 |
| DUSP11 | BNIPL | 0.692483 | 3 | 0 | 3 |
| DUSP11 | CYB5R2 | 0.687498 | 5 | 0 | 5 |
| DUSP11 | KRT6A | 0.677937 | 6 | 0 | 5 |
| DUSP11 | ZNF710 | 0.676125 | 3 | 0 | 3 |
| DUSP11 | LIPH | 0.669853 | 3 | 0 | 3 |
| DUSP11 | ZMAT2 | 0.663524 | 3 | 0 | 3 |
| DUSP11 | DUOXA1 | 0.663373 | 4 | 0 | 4 |
| DUSP11 | TGFA | 0.662759 | 6 | 0 | 5 |
| DUSP11 | SMIM15 | 0.661495 | 3 | 0 | 3 |
| DUSP11 | TUBB3 | 0.661203 | 3 | 0 | 3 |
| DUSP11 | CMAS | 0.658187 | 8 | 0 | 7 |
| DUSP11 | ACBD3 | 0.656553 | 6 | 0 | 5 |
| DUSP11 | CTTNBP2NL | 0.654031 | 4 | 0 | 3 |
| DUSP11 | CNIH1 | 0.653453 | 4 | 0 | 4 |
For details and further investigation, click here