| Full name: cytidine monophosphate N-acetylneuraminic acid synthetase | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 12p12.1 | ||
| Entrez ID: 55907 | HGNC ID: HGNC:18290 | Ensembl Gene: ENSG00000111726 | OMIM ID: 603316 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of CMAS:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CMAS | 55907 | 218111_s_at | -0.7601 | 0.2347 | |
| GSE20347 | CMAS | 55907 | 218111_s_at | -1.1376 | 0.0002 | |
| GSE23400 | CMAS | 55907 | 218111_s_at | -0.8578 | 0.0000 | |
| GSE26886 | CMAS | 55907 | 218111_s_at | -2.1278 | 0.0000 | |
| GSE29001 | CMAS | 55907 | 218111_s_at | -0.8058 | 0.0926 | |
| GSE38129 | CMAS | 55907 | 218111_s_at | -0.5079 | 0.1087 | |
| GSE45670 | CMAS | 55907 | 218111_s_at | -0.5855 | 0.0054 | |
| GSE53622 | CMAS | 55907 | 52667 | -0.8022 | 0.0000 | |
| GSE53624 | CMAS | 55907 | 14278 | -1.1709 | 0.0000 | |
| GSE63941 | CMAS | 55907 | 218111_s_at | 0.5705 | 0.2586 | |
| GSE77861 | CMAS | 55907 | 218111_s_at | -0.8267 | 0.0476 | |
| GSE97050 | CMAS | 55907 | A_32_P73045 | -0.3525 | 0.2569 | |
| SRP007169 | CMAS | 55907 | RNAseq | -1.6680 | 0.0000 | |
| SRP008496 | CMAS | 55907 | RNAseq | -1.1785 | 0.0005 | |
| SRP064894 | CMAS | 55907 | RNAseq | -1.3019 | 0.0000 | |
| SRP133303 | CMAS | 55907 | RNAseq | -0.7578 | 0.0007 | |
| SRP159526 | CMAS | 55907 | RNAseq | -1.3115 | 0.0000 | |
| SRP193095 | CMAS | 55907 | RNAseq | -1.6267 | 0.0000 | |
| SRP219564 | CMAS | 55907 | RNAseq | -1.4096 | 0.0026 | |
| TCGA | CMAS | 55907 | RNAseq | 0.1301 | 0.0322 |
Upregulated datasets: 0; Downregulated datasets: 9.
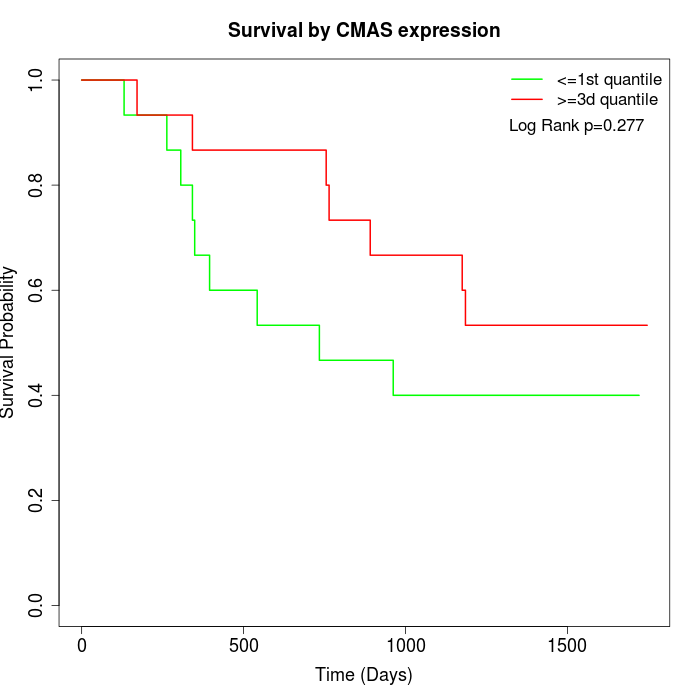
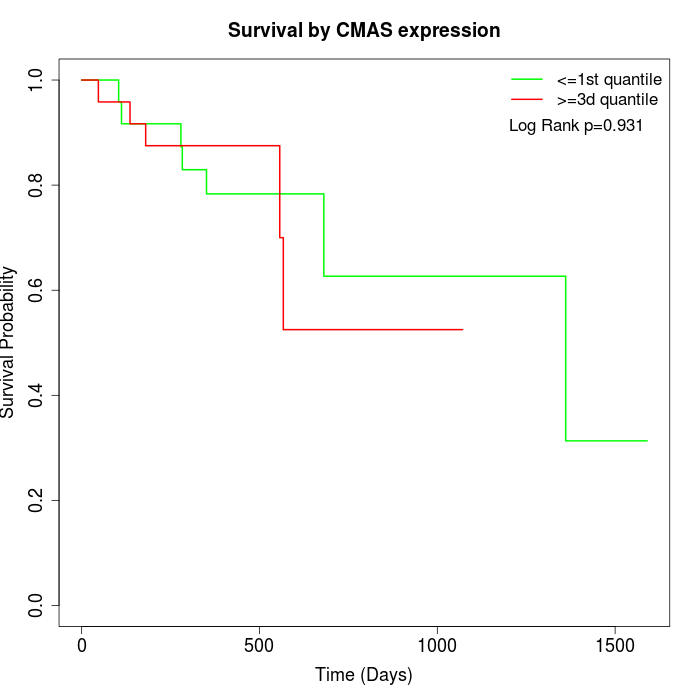
Survival by CMAS expression:
Note: Click image to view full size file.
Copy number change of CMAS:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CMAS | 55907 | 10 | 1 | 19 | |
| GSE20123 | CMAS | 55907 | 10 | 1 | 19 | |
| GSE43470 | CMAS | 55907 | 9 | 4 | 30 | |
| GSE46452 | CMAS | 55907 | 10 | 1 | 48 | |
| GSE47630 | CMAS | 55907 | 14 | 1 | 25 | |
| GSE54993 | CMAS | 55907 | 1 | 9 | 60 | |
| GSE54994 | CMAS | 55907 | 11 | 2 | 40 | |
| GSE60625 | CMAS | 55907 | 0 | 1 | 10 | |
| GSE74703 | CMAS | 55907 | 9 | 3 | 24 | |
| GSE74704 | CMAS | 55907 | 6 | 1 | 13 | |
| TCGA | CMAS | 55907 | 43 | 5 | 48 |
Total number of gains: 123; Total number of losses: 29; Total Number of normals: 336.
Somatic mutations of CMAS:
Generating mutation plots.
Highly correlated genes for CMAS:
Showing top 20/1266 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CMAS | TAB3 | 0.764285 | 6 | 0 | 6 |
| CMAS | A2ML1 | 0.736707 | 7 | 0 | 6 |
| CMAS | CNFN | 0.736449 | 6 | 0 | 6 |
| CMAS | SPNS2 | 0.734421 | 4 | 0 | 4 |
| CMAS | C15orf48 | 0.734245 | 6 | 0 | 5 |
| CMAS | SH3PXD2A-AS1 | 0.733931 | 5 | 0 | 5 |
| CMAS | VSIG2 | 0.733424 | 6 | 0 | 6 |
| CMAS | EPGN | 0.730081 | 3 | 0 | 3 |
| CMAS | RAET1E | 0.72801 | 6 | 0 | 6 |
| CMAS | ATP6V1C2 | 0.721695 | 4 | 0 | 4 |
| CMAS | FAM3D | 0.719999 | 7 | 0 | 7 |
| CMAS | LNX1 | 0.717025 | 6 | 0 | 6 |
| CMAS | HCG22 | 0.716917 | 6 | 0 | 6 |
| CMAS | ATG9B | 0.716826 | 7 | 0 | 7 |
| CMAS | GYS2 | 0.71568 | 10 | 0 | 10 |
| CMAS | SH3GL1 | 0.711641 | 9 | 0 | 9 |
| CMAS | C4orf3 | 0.707945 | 7 | 0 | 7 |
| CMAS | NANOS2 | 0.707703 | 3 | 0 | 3 |
| CMAS | ZDHHC20 | 0.706071 | 3 | 0 | 3 |
| CMAS | FAM214A | 0.704379 | 6 | 0 | 6 |
For details and further investigation, click here