| Full name: zinc finger matrin-type 2 | Alias Symbol: FLJ31121|hSNU23|Snu23 | ||
| Type: protein-coding gene | Cytoband: 5q31.3 | ||
| Entrez ID: 153527 | HGNC ID: HGNC:26433 | Ensembl Gene: ENSG00000146007 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of ZMAT2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ZMAT2 | 153527 | 224782_at | -0.5848 | 0.1199 | |
| GSE26886 | ZMAT2 | 153527 | 224782_at | -1.3859 | 0.0000 | |
| GSE45670 | ZMAT2 | 153527 | 224782_at | -0.1103 | 0.4158 | |
| GSE53622 | ZMAT2 | 153527 | 37967 | -0.2298 | 0.0001 | |
| GSE53624 | ZMAT2 | 153527 | 37967 | -0.3073 | 0.0000 | |
| GSE63941 | ZMAT2 | 153527 | 224782_at | 0.5928 | 0.1324 | |
| GSE77861 | ZMAT2 | 153527 | 224782_at | -0.3325 | 0.3453 | |
| GSE97050 | ZMAT2 | 153527 | A_23_P346302 | 0.0434 | 0.8370 | |
| SRP007169 | ZMAT2 | 153527 | RNAseq | -0.5702 | 0.2871 | |
| SRP008496 | ZMAT2 | 153527 | RNAseq | -0.6977 | 0.0337 | |
| SRP064894 | ZMAT2 | 153527 | RNAseq | -0.3836 | 0.0352 | |
| SRP133303 | ZMAT2 | 153527 | RNAseq | 0.1794 | 0.4655 | |
| SRP159526 | ZMAT2 | 153527 | RNAseq | -0.3564 | 0.3255 | |
| SRP193095 | ZMAT2 | 153527 | RNAseq | -0.6869 | 0.0000 | |
| SRP219564 | ZMAT2 | 153527 | RNAseq | -0.4412 | 0.0846 | |
| TCGA | ZMAT2 | 153527 | RNAseq | 0.0216 | 0.6657 |
Upregulated datasets: 0; Downregulated datasets: 1.
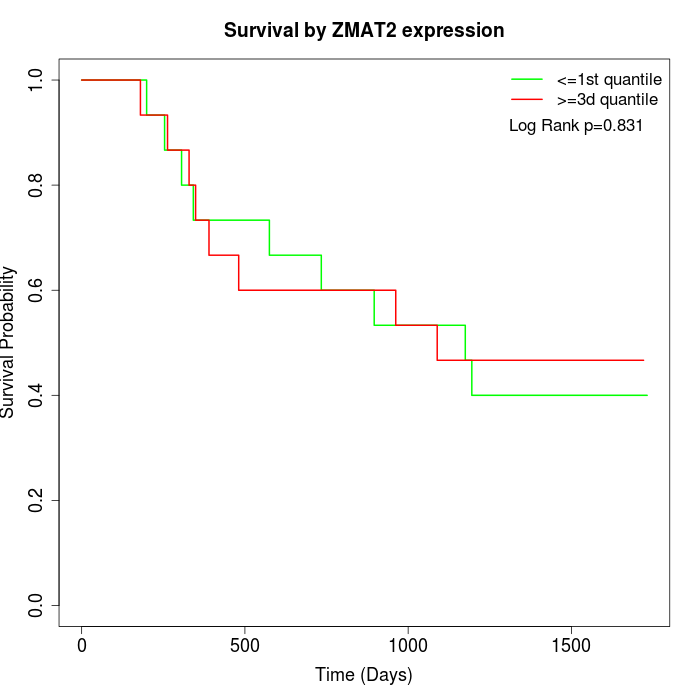
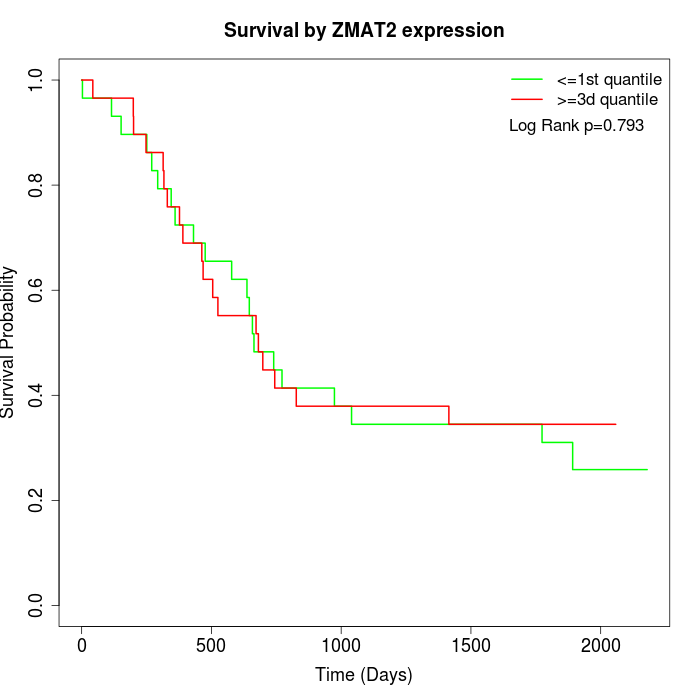
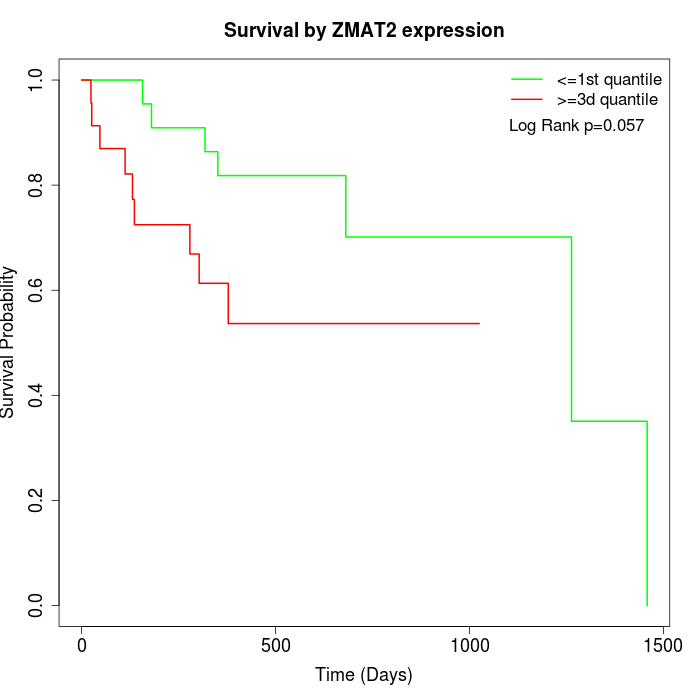
Survival by ZMAT2 expression:
Note: Click image to view full size file.
Copy number change of ZMAT2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ZMAT2 | 153527 | 3 | 11 | 16 | |
| GSE20123 | ZMAT2 | 153527 | 4 | 11 | 15 | |
| GSE43470 | ZMAT2 | 153527 | 2 | 10 | 31 | |
| GSE46452 | ZMAT2 | 153527 | 0 | 27 | 32 | |
| GSE47630 | ZMAT2 | 153527 | 0 | 20 | 20 | |
| GSE54993 | ZMAT2 | 153527 | 9 | 1 | 60 | |
| GSE54994 | ZMAT2 | 153527 | 2 | 14 | 37 | |
| GSE60625 | ZMAT2 | 153527 | 0 | 0 | 11 | |
| GSE74703 | ZMAT2 | 153527 | 2 | 7 | 27 | |
| GSE74704 | ZMAT2 | 153527 | 3 | 5 | 12 | |
| TCGA | ZMAT2 | 153527 | 4 | 37 | 55 |
Total number of gains: 29; Total number of losses: 143; Total Number of normals: 316.
Somatic mutations of ZMAT2:
Generating mutation plots.
Highly correlated genes for ZMAT2:
Showing top 20/585 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ZMAT2 | KDM6A | 0.754531 | 3 | 0 | 3 |
| ZMAT2 | VDAC1 | 0.731748 | 5 | 0 | 5 |
| ZMAT2 | MTMR3 | 0.717829 | 3 | 0 | 3 |
| ZMAT2 | UBE2D2 | 0.716371 | 7 | 0 | 6 |
| ZMAT2 | TNKS2 | 0.71363 | 3 | 0 | 3 |
| ZMAT2 | CPT2 | 0.710976 | 4 | 0 | 4 |
| ZMAT2 | AFF4 | 0.703571 | 5 | 0 | 5 |
| ZMAT2 | SLC35F5 | 0.701286 | 3 | 0 | 3 |
| ZMAT2 | SCYL2 | 0.687393 | 3 | 0 | 3 |
| ZMAT2 | HBP1 | 0.683844 | 5 | 0 | 4 |
| ZMAT2 | ACAD8 | 0.677977 | 4 | 0 | 3 |
| ZMAT2 | MBOAT2 | 0.673424 | 4 | 0 | 4 |
| ZMAT2 | CAPNS2 | 0.671978 | 5 | 0 | 4 |
| ZMAT2 | SEC14L5 | 0.671727 | 3 | 0 | 3 |
| ZMAT2 | GPX3 | 0.669867 | 6 | 0 | 4 |
| ZMAT2 | ETF1 | 0.667937 | 6 | 0 | 5 |
| ZMAT2 | CRCT1 | 0.6679 | 5 | 0 | 4 |
| ZMAT2 | IK | 0.66778 | 8 | 0 | 7 |
| ZMAT2 | SAR1B | 0.667529 | 7 | 0 | 6 |
| ZMAT2 | DUSP11 | 0.663524 | 3 | 0 | 3 |
For details and further investigation, click here