| Full name: dual specificity phosphatase 14 | Alias Symbol: MKP-L|MKP6 | ||
| Type: protein-coding gene | Cytoband: 17q12 | ||
| Entrez ID: 11072 | HGNC ID: HGNC:17007 | Ensembl Gene: ENSG00000276023 | OMIM ID: 606618 |
| Drug and gene relationship at DGIdb | |||
Expression of DUSP14:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | DUSP14 | 11072 | 203367_at | 0.5334 | 0.4578 | |
| GSE20347 | DUSP14 | 11072 | 203367_at | -0.1996 | 0.5113 | |
| GSE23400 | DUSP14 | 11072 | 203367_at | -0.0716 | 0.5388 | |
| GSE26886 | DUSP14 | 11072 | 203367_at | 0.5925 | 0.1252 | |
| GSE29001 | DUSP14 | 11072 | 203367_at | -0.6010 | 0.1012 | |
| GSE38129 | DUSP14 | 11072 | 203367_at | 0.0585 | 0.8434 | |
| GSE45670 | DUSP14 | 11072 | 203367_at | -0.3108 | 0.3406 | |
| GSE53622 | DUSP14 | 11072 | 10950 | 0.1131 | 0.4749 | |
| GSE53624 | DUSP14 | 11072 | 10950 | -0.2874 | 0.0091 | |
| GSE63941 | DUSP14 | 11072 | 203367_at | -0.7017 | 0.2148 | |
| GSE77861 | DUSP14 | 11072 | 203367_at | 0.2602 | 0.6321 | |
| GSE97050 | DUSP14 | 11072 | A_23_P207537 | -0.3570 | 0.5139 | |
| SRP007169 | DUSP14 | 11072 | RNAseq | -2.4816 | 0.0000 | |
| SRP008496 | DUSP14 | 11072 | RNAseq | -1.7908 | 0.0001 | |
| SRP064894 | DUSP14 | 11072 | RNAseq | -0.9820 | 0.0012 | |
| SRP133303 | DUSP14 | 11072 | RNAseq | -0.0008 | 0.9980 | |
| SRP159526 | DUSP14 | 11072 | RNAseq | -0.2199 | 0.6369 | |
| SRP193095 | DUSP14 | 11072 | RNAseq | -0.0237 | 0.9402 | |
| SRP219564 | DUSP14 | 11072 | RNAseq | -0.7541 | 0.0759 | |
| TCGA | DUSP14 | 11072 | RNAseq | 0.4331 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 2.
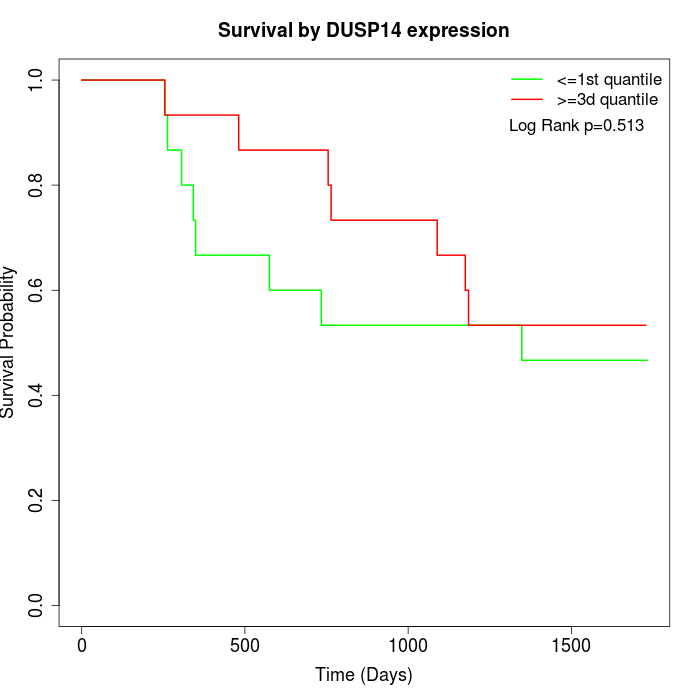
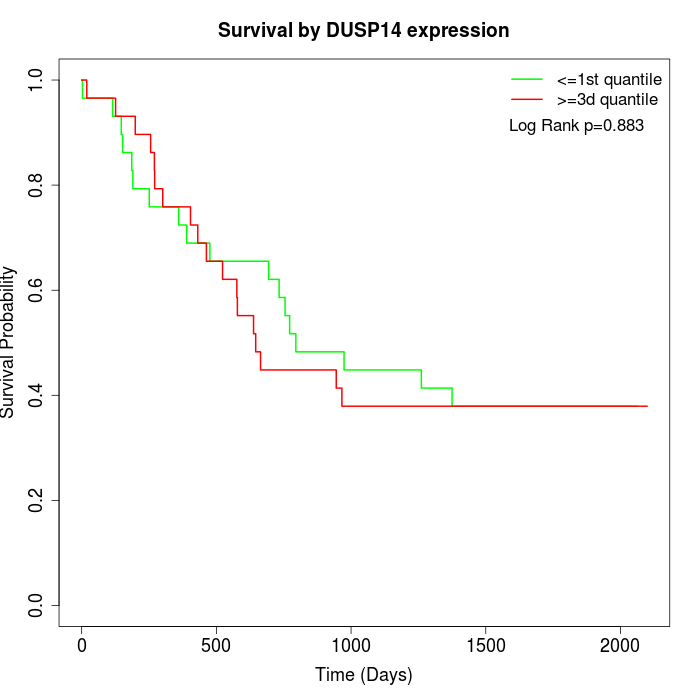
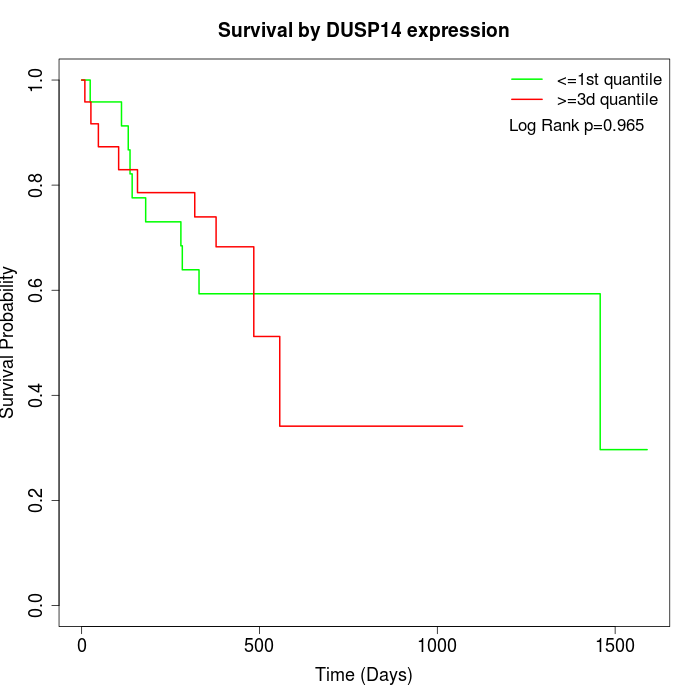
Survival by DUSP14 expression:
Note: Click image to view full size file.
Copy number change of DUSP14:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | DUSP14 | 11072 | 6 | 1 | 23 | |
| GSE20123 | DUSP14 | 11072 | 6 | 1 | 23 | |
| GSE43470 | DUSP14 | 11072 | 1 | 2 | 40 | |
| GSE46452 | DUSP14 | 11072 | 34 | 0 | 25 | |
| GSE47630 | DUSP14 | 11072 | 8 | 1 | 31 | |
| GSE54993 | DUSP14 | 11072 | 3 | 3 | 64 | |
| GSE54994 | DUSP14 | 11072 | 9 | 5 | 39 | |
| GSE60625 | DUSP14 | 11072 | 4 | 0 | 7 | |
| GSE74703 | DUSP14 | 11072 | 1 | 1 | 34 | |
| GSE74704 | DUSP14 | 11072 | 3 | 1 | 16 | |
| TCGA | DUSP14 | 11072 | 19 | 10 | 67 |
Total number of gains: 94; Total number of losses: 25; Total Number of normals: 369.
Somatic mutations of DUSP14:
Generating mutation plots.
Highly correlated genes for DUSP14:
Showing top 20/87 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| DUSP14 | TNPO1 | 0.681353 | 3 | 0 | 3 |
| DUSP14 | C16orf87 | 0.677695 | 4 | 0 | 4 |
| DUSP14 | ERCC4 | 0.670023 | 3 | 0 | 3 |
| DUSP14 | C12orf75 | 0.643746 | 3 | 0 | 3 |
| DUSP14 | INO80D | 0.642846 | 3 | 0 | 3 |
| DUSP14 | WDR41 | 0.641883 | 3 | 0 | 3 |
| DUSP14 | KCNG2 | 0.616663 | 4 | 0 | 3 |
| DUSP14 | N4BP1 | 0.613285 | 5 | 0 | 4 |
| DUSP14 | PSMD12 | 0.596684 | 5 | 0 | 3 |
| DUSP14 | RAB30 | 0.596603 | 3 | 0 | 3 |
| DUSP14 | GNG12 | 0.595385 | 4 | 0 | 3 |
| DUSP14 | SLC19A2 | 0.583719 | 4 | 0 | 3 |
| DUSP14 | VEZT | 0.578277 | 4 | 0 | 3 |
| DUSP14 | XPO6 | 0.57581 | 4 | 0 | 3 |
| DUSP14 | WFDC12 | 0.575483 | 5 | 0 | 4 |
| DUSP14 | SLC31A2 | 0.575351 | 6 | 0 | 5 |
| DUSP14 | TRMT6 | 0.573047 | 3 | 0 | 3 |
| DUSP14 | CHORDC1 | 0.57286 | 5 | 0 | 3 |
| DUSP14 | FAM49A | 0.572049 | 5 | 0 | 3 |
| DUSP14 | DIP2B | 0.571449 | 6 | 0 | 4 |
For details and further investigation, click here