| Full name: unconventional SNARE in the ER 1 | Alias Symbol: p31|SLT1|MDS032|D12 | ||
| Type: protein-coding gene | Cytoband: 19p13.11 | ||
| Entrez ID: 55850 | HGNC ID: HGNC:30882 | Ensembl Gene: ENSG00000053501 | OMIM ID: 610675 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of USE1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | USE1 | 55850 | 221706_s_at | 0.1912 | 0.6789 | |
| GSE20347 | USE1 | 55850 | 219348_at | -0.0990 | 0.4430 | |
| GSE23400 | USE1 | 55850 | 221706_s_at | 0.1115 | 0.0347 | |
| GSE26886 | USE1 | 55850 | 219348_at | 0.2282 | 0.2380 | |
| GSE29001 | USE1 | 55850 | 219348_at | 0.5309 | 0.1904 | |
| GSE38129 | USE1 | 55850 | 219348_at | -0.0164 | 0.9180 | |
| GSE45670 | USE1 | 55850 | 219348_at | 0.0755 | 0.4788 | |
| GSE53622 | USE1 | 55850 | 5732 | 0.0924 | 0.1452 | |
| GSE53624 | USE1 | 55850 | 5732 | 0.2468 | 0.0002 | |
| GSE63941 | USE1 | 55850 | 219348_at | -0.7813 | 0.1105 | |
| GSE77861 | USE1 | 55850 | 219348_at | -0.0975 | 0.3466 | |
| GSE97050 | USE1 | 55850 | A_23_P79122 | 0.0722 | 0.8278 | |
| SRP007169 | USE1 | 55850 | RNAseq | 0.3258 | 0.6440 | |
| SRP064894 | USE1 | 55850 | RNAseq | 0.5382 | 0.1192 | |
| SRP133303 | USE1 | 55850 | RNAseq | -0.1799 | 0.3143 | |
| SRP159526 | USE1 | 55850 | RNAseq | 0.2068 | 0.3262 | |
| SRP193095 | USE1 | 55850 | RNAseq | 0.1105 | 0.3869 | |
| SRP219564 | USE1 | 55850 | RNAseq | 0.3589 | 0.1348 | |
| TCGA | USE1 | 55850 | RNAseq | 0.0926 | 0.2296 |
Upregulated datasets: 0; Downregulated datasets: 0.
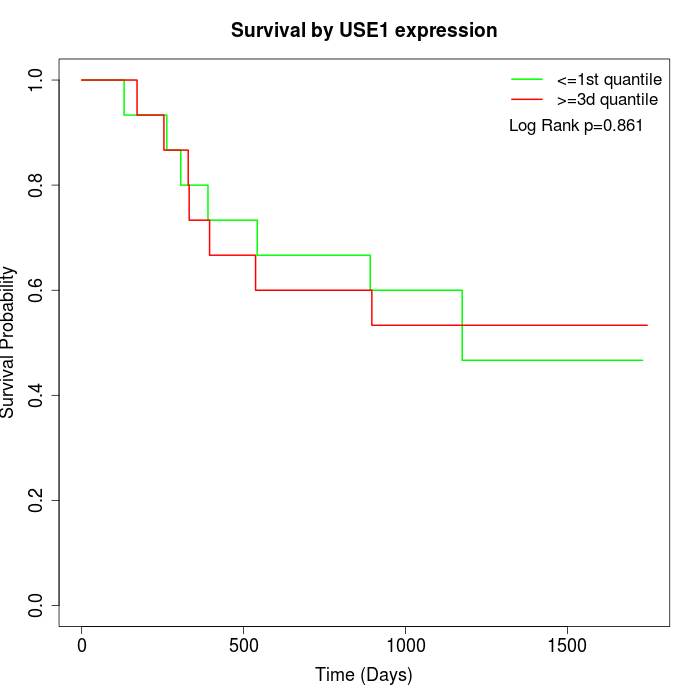
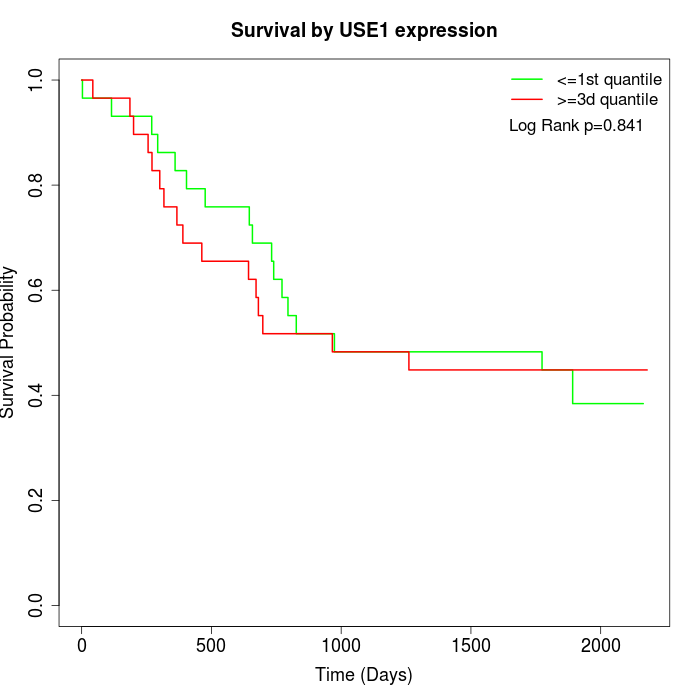
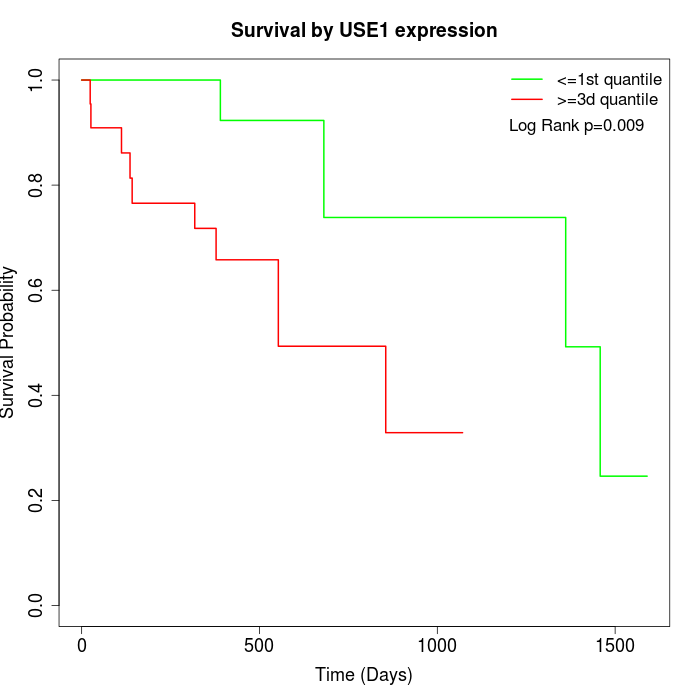
Survival by USE1 expression:
Note: Click image to view full size file.
Copy number change of USE1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | USE1 | 55850 | 4 | 4 | 22 | |
| GSE20123 | USE1 | 55850 | 3 | 2 | 25 | |
| GSE43470 | USE1 | 55850 | 2 | 6 | 35 | |
| GSE46452 | USE1 | 55850 | 47 | 1 | 11 | |
| GSE47630 | USE1 | 55850 | 4 | 8 | 28 | |
| GSE54993 | USE1 | 55850 | 15 | 4 | 51 | |
| GSE54994 | USE1 | 55850 | 6 | 14 | 33 | |
| GSE60625 | USE1 | 55850 | 9 | 0 | 2 | |
| GSE74703 | USE1 | 55850 | 2 | 4 | 30 | |
| GSE74704 | USE1 | 55850 | 0 | 2 | 18 | |
| TCGA | USE1 | 55850 | 17 | 9 | 70 |
Total number of gains: 109; Total number of losses: 54; Total Number of normals: 325.
Somatic mutations of USE1:
Generating mutation plots.
Highly correlated genes for USE1:
Showing top 20/338 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| USE1 | THOC5 | 0.792363 | 3 | 0 | 3 |
| USE1 | C9orf78 | 0.791666 | 3 | 0 | 3 |
| USE1 | AZIN1 | 0.77953 | 3 | 0 | 3 |
| USE1 | NASP | 0.770897 | 3 | 0 | 3 |
| USE1 | SNRPC | 0.746341 | 3 | 0 | 3 |
| USE1 | ATG4C | 0.73901 | 3 | 0 | 3 |
| USE1 | NSUN3 | 0.738559 | 3 | 0 | 3 |
| USE1 | ADNP | 0.735484 | 3 | 0 | 3 |
| USE1 | CTSZ | 0.731967 | 3 | 0 | 3 |
| USE1 | RAD18 | 0.727292 | 3 | 0 | 3 |
| USE1 | RING1 | 0.721768 | 3 | 0 | 3 |
| USE1 | ZNF616 | 0.718736 | 3 | 0 | 3 |
| USE1 | PNO1 | 0.714442 | 3 | 0 | 3 |
| USE1 | ZNF3 | 0.714358 | 4 | 0 | 3 |
| USE1 | ZNF628 | 0.710281 | 4 | 0 | 4 |
| USE1 | GTPBP6 | 0.707768 | 4 | 0 | 3 |
| USE1 | ZNF35 | 0.70299 | 3 | 0 | 3 |
| USE1 | SFXN2 | 0.702505 | 3 | 0 | 3 |
| USE1 | FTO | 0.698837 | 3 | 0 | 3 |
| USE1 | ECHS1 | 0.695352 | 3 | 0 | 3 |
For details and further investigation, click here