| Full name: Ras related GTP binding A | Alias Symbol: RAGA|FIP-1 | ||
| Type: protein-coding gene | Cytoband: 9p22.1 | ||
| Entrez ID: 10670 | HGNC ID: HGNC:16963 | Ensembl Gene: ENSG00000155876 | OMIM ID: 612194 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
RRAGA involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04150 | mTOR signaling pathway |
Expression of RRAGA:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RRAGA | 10670 | 201628_s_at | -0.5353 | 0.1194 | |
| GSE20347 | RRAGA | 10670 | 201628_s_at | -0.5724 | 0.0013 | |
| GSE23400 | RRAGA | 10670 | 201628_s_at | -0.0253 | 0.6627 | |
| GSE26886 | RRAGA | 10670 | 201628_s_at | -0.4212 | 0.0266 | |
| GSE29001 | RRAGA | 10670 | 201628_s_at | -0.1285 | 0.6318 | |
| GSE38129 | RRAGA | 10670 | 201628_s_at | -0.3933 | 0.0058 | |
| GSE45670 | RRAGA | 10670 | 201628_s_at | -0.3425 | 0.0118 | |
| GSE53622 | RRAGA | 10670 | 17420 | -0.4822 | 0.0000 | |
| GSE53624 | RRAGA | 10670 | 17420 | -0.1030 | 0.0950 | |
| GSE63941 | RRAGA | 10670 | 201628_s_at | -1.0762 | 0.1249 | |
| GSE77861 | RRAGA | 10670 | 201628_s_at | -0.4755 | 0.0411 | |
| GSE97050 | RRAGA | 10670 | A_23_P169117 | -0.3009 | 0.1836 | |
| SRP007169 | RRAGA | 10670 | RNAseq | -0.8804 | 0.0206 | |
| SRP008496 | RRAGA | 10670 | RNAseq | -0.6724 | 0.0115 | |
| SRP064894 | RRAGA | 10670 | RNAseq | 0.0029 | 0.9839 | |
| SRP133303 | RRAGA | 10670 | RNAseq | -0.2441 | 0.0917 | |
| SRP159526 | RRAGA | 10670 | RNAseq | -0.5896 | 0.0116 | |
| SRP193095 | RRAGA | 10670 | RNAseq | -0.5918 | 0.0000 | |
| SRP219564 | RRAGA | 10670 | RNAseq | -0.4245 | 0.2184 | |
| TCGA | RRAGA | 10670 | RNAseq | -0.0339 | 0.4891 |
Upregulated datasets: 0; Downregulated datasets: 0.
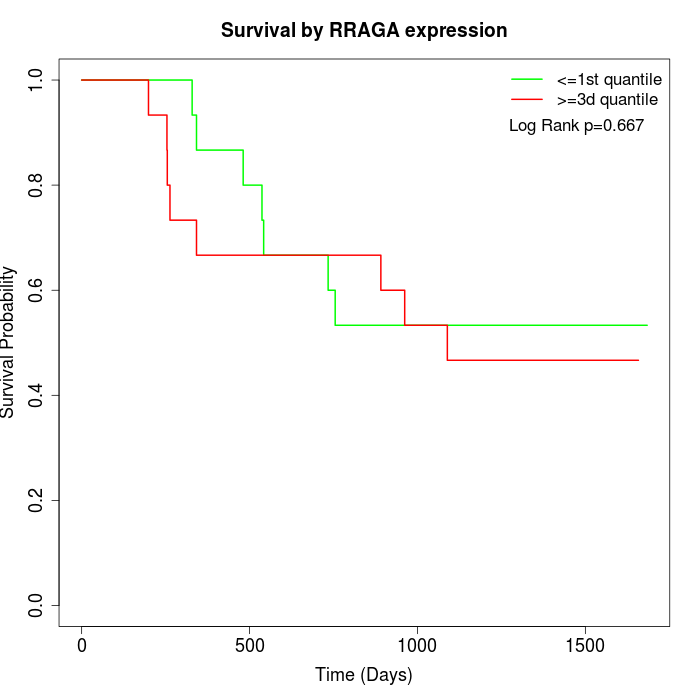
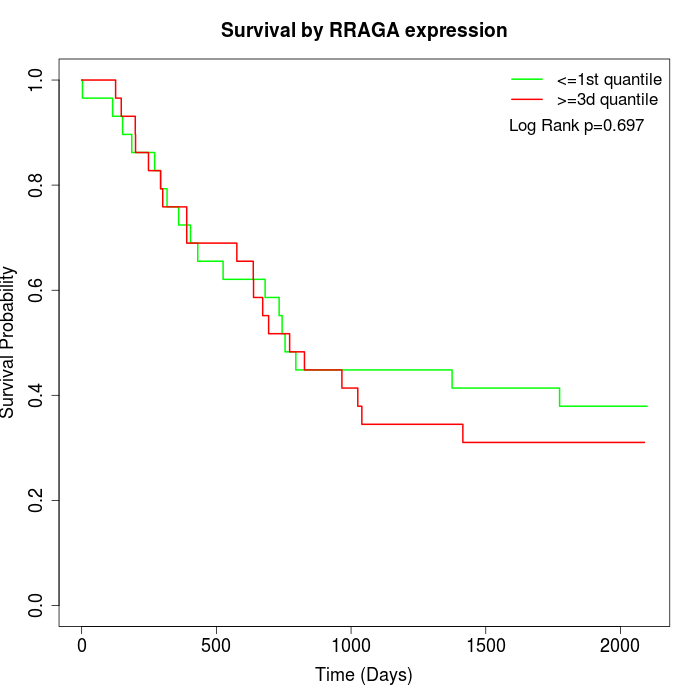
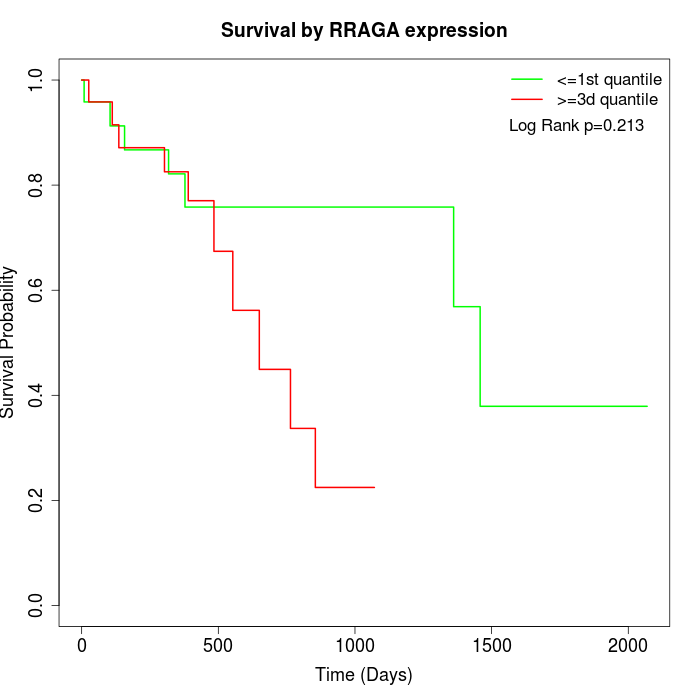
Survival by RRAGA expression:
Note: Click image to view full size file.
Copy number change of RRAGA:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RRAGA | 10670 | 2 | 16 | 12 | |
| GSE20123 | RRAGA | 10670 | 2 | 16 | 12 | |
| GSE43470 | RRAGA | 10670 | 5 | 11 | 27 | |
| GSE46452 | RRAGA | 10670 | 2 | 23 | 34 | |
| GSE47630 | RRAGA | 10670 | 3 | 27 | 10 | |
| GSE54993 | RRAGA | 10670 | 6 | 2 | 62 | |
| GSE54994 | RRAGA | 10670 | 9 | 16 | 28 | |
| GSE60625 | RRAGA | 10670 | 0 | 0 | 11 | |
| GSE74703 | RRAGA | 10670 | 5 | 8 | 23 | |
| GSE74704 | RRAGA | 10670 | 0 | 13 | 7 | |
| TCGA | RRAGA | 10670 | 12 | 51 | 33 |
Total number of gains: 46; Total number of losses: 183; Total Number of normals: 259.
Somatic mutations of RRAGA:
Generating mutation plots.
Highly correlated genes for RRAGA:
Showing top 20/1004 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RRAGA | TLE2 | 0.824209 | 3 | 0 | 3 |
| RRAGA | SNX12 | 0.810269 | 3 | 0 | 3 |
| RRAGA | ATG2B | 0.778247 | 3 | 0 | 3 |
| RRAGA | TMEM139 | 0.769119 | 3 | 0 | 3 |
| RRAGA | ECSCR | 0.767784 | 3 | 0 | 3 |
| RRAGA | COG6 | 0.755229 | 3 | 0 | 3 |
| RRAGA | SNX25 | 0.754608 | 3 | 0 | 3 |
| RRAGA | AIFM1 | 0.739501 | 3 | 0 | 3 |
| RRAGA | THAP8 | 0.731452 | 3 | 0 | 3 |
| RRAGA | UBR2 | 0.725 | 5 | 0 | 4 |
| RRAGA | RBM5 | 0.724902 | 4 | 0 | 4 |
| RRAGA | FBXO10 | 0.723316 | 4 | 0 | 4 |
| RRAGA | RORA | 0.716972 | 6 | 0 | 6 |
| RRAGA | RIC3 | 0.714003 | 4 | 0 | 4 |
| RRAGA | UCHL3 | 0.713649 | 5 | 0 | 5 |
| RRAGA | HLCS | 0.707784 | 3 | 0 | 3 |
| RRAGA | SLC22A15 | 0.703641 | 4 | 0 | 4 |
| RRAGA | TMEM41B | 0.703392 | 3 | 0 | 3 |
| RRAGA | MAGEC1 | 0.701807 | 3 | 0 | 3 |
| RRAGA | AK3 | 0.698431 | 8 | 0 | 7 |
For details and further investigation, click here