| Full name: endothelin 1 | Alias Symbol: ET1 | ||
| Type: protein-coding gene | Cytoband: 6p24.1 | ||
| Entrez ID: 1906 | HGNC ID: HGNC:3176 | Ensembl Gene: ENSG00000078401 | OMIM ID: 131240 |
| Drug and gene relationship at DGIdb | |||
EDN1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04066 | HIF-1 signaling pathway | |
| hsa04916 | Melanogenesis | |
| hsa04933 | AGE-RAGE signaling pathway in diabetic complications |
Expression of EDN1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | EDN1 | 1906 | 222802_at | -0.9650 | 0.0763 | |
| GSE20347 | EDN1 | 1906 | 218995_s_at | 0.4345 | 0.1332 | |
| GSE23400 | EDN1 | 1906 | 218995_s_at | 0.4453 | 0.0001 | |
| GSE26886 | EDN1 | 1906 | 222802_at | 1.1265 | 0.0027 | |
| GSE29001 | EDN1 | 1906 | 218995_s_at | 0.0278 | 0.9535 | |
| GSE38129 | EDN1 | 1906 | 218995_s_at | 0.2864 | 0.2841 | |
| GSE45670 | EDN1 | 1906 | 218995_s_at | -0.3959 | 0.3503 | |
| GSE53622 | EDN1 | 1906 | 140 | -0.1987 | 0.2133 | |
| GSE53624 | EDN1 | 1906 | 140 | 0.1970 | 0.1053 | |
| GSE63941 | EDN1 | 1906 | 222802_at | -1.5206 | 0.2578 | |
| GSE77861 | EDN1 | 1906 | 222802_at | 1.0324 | 0.0179 | |
| GSE97050 | EDN1 | 1906 | A_23_P214821 | 0.7381 | 0.2151 | |
| SRP007169 | EDN1 | 1906 | RNAseq | 1.3974 | 0.0188 | |
| SRP064894 | EDN1 | 1906 | RNAseq | 0.0332 | 0.9376 | |
| SRP133303 | EDN1 | 1906 | RNAseq | 0.4297 | 0.3250 | |
| SRP159526 | EDN1 | 1906 | RNAseq | 0.6243 | 0.2134 | |
| SRP193095 | EDN1 | 1906 | RNAseq | 2.0997 | 0.0000 | |
| SRP219564 | EDN1 | 1906 | RNAseq | -0.0110 | 0.9861 | |
| TCGA | EDN1 | 1906 | RNAseq | -0.4034 | 0.0027 |
Upregulated datasets: 4; Downregulated datasets: 0.
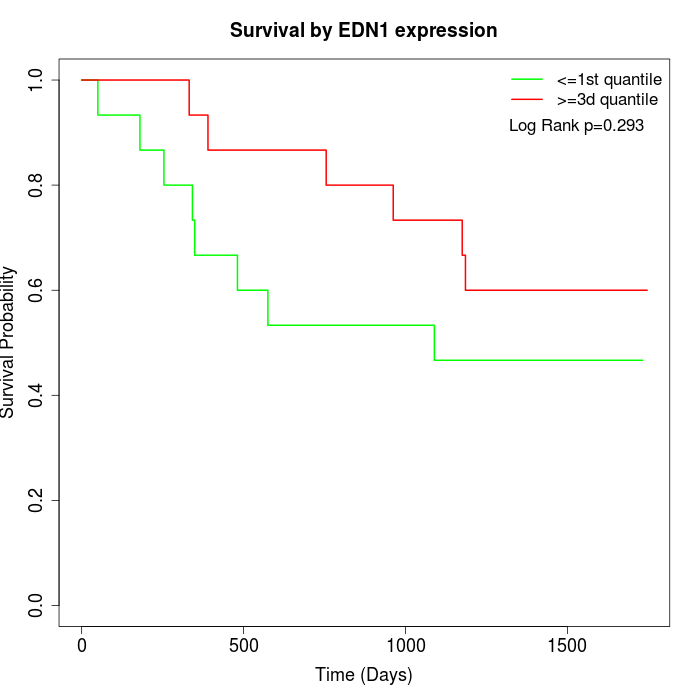
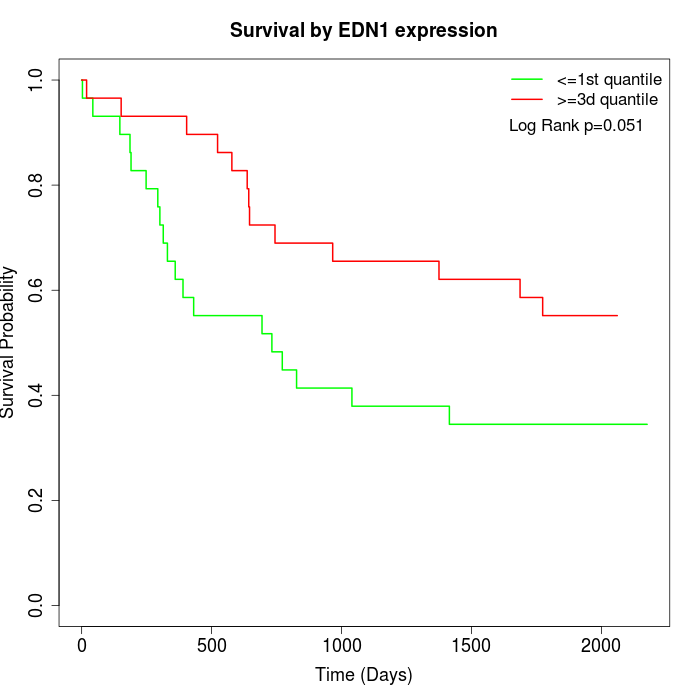
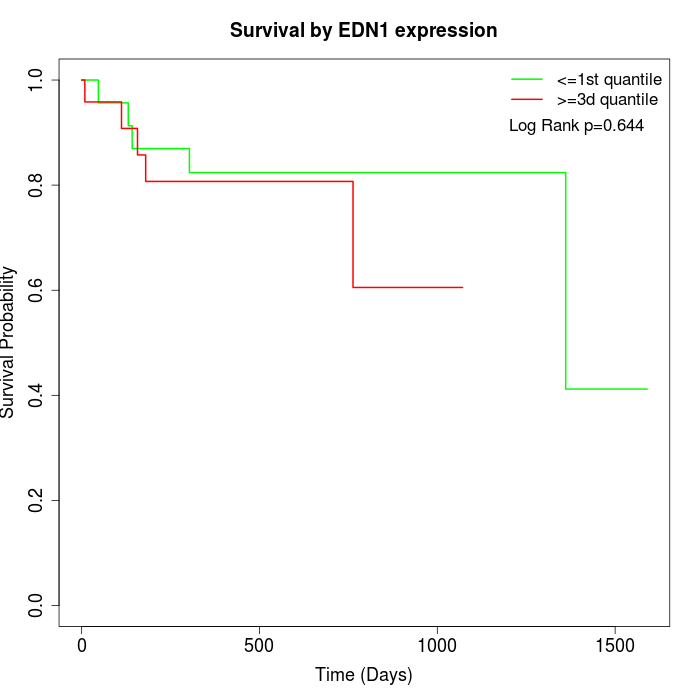
Survival by EDN1 expression:
Note: Click image to view full size file.
Copy number change of EDN1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | EDN1 | 1906 | 4 | 7 | 19 | |
| GSE20123 | EDN1 | 1906 | 4 | 7 | 19 | |
| GSE43470 | EDN1 | 1906 | 8 | 1 | 34 | |
| GSE46452 | EDN1 | 1906 | 1 | 10 | 48 | |
| GSE47630 | EDN1 | 1906 | 7 | 8 | 25 | |
| GSE54993 | EDN1 | 1906 | 1 | 1 | 68 | |
| GSE54994 | EDN1 | 1906 | 10 | 3 | 40 | |
| GSE60625 | EDN1 | 1906 | 0 | 1 | 10 | |
| GSE74703 | EDN1 | 1906 | 7 | 1 | 28 | |
| GSE74704 | EDN1 | 1906 | 2 | 3 | 15 | |
| TCGA | EDN1 | 1906 | 16 | 26 | 54 |
Total number of gains: 60; Total number of losses: 68; Total Number of normals: 360.
Somatic mutations of EDN1:
Generating mutation plots.
Highly correlated genes for EDN1:
Showing top 20/241 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| EDN1 | ARTN | 0.739008 | 3 | 0 | 3 |
| EDN1 | FBLN1 | 0.737 | 3 | 0 | 3 |
| EDN1 | MARVELD1 | 0.719331 | 3 | 0 | 3 |
| EDN1 | SLC6A6 | 0.700345 | 3 | 0 | 3 |
| EDN1 | CNTNAP1 | 0.694679 | 3 | 0 | 3 |
| EDN1 | USP5 | 0.679128 | 3 | 0 | 3 |
| EDN1 | ABHD1 | 0.676817 | 3 | 0 | 3 |
| EDN1 | CD151 | 0.676805 | 3 | 0 | 3 |
| EDN1 | WDR54 | 0.67185 | 4 | 0 | 3 |
| EDN1 | TPP1 | 0.671655 | 3 | 0 | 3 |
| EDN1 | PHLDB2 | 0.671229 | 4 | 0 | 3 |
| EDN1 | MED27 | 0.668691 | 3 | 0 | 3 |
| EDN1 | SEL1L3 | 0.665215 | 3 | 0 | 3 |
| EDN1 | S1PR1 | 0.662474 | 4 | 0 | 3 |
| EDN1 | KLF7 | 0.655817 | 5 | 0 | 5 |
| EDN1 | SYNPO | 0.65522 | 3 | 0 | 3 |
| EDN1 | KLHL13 | 0.653885 | 4 | 0 | 3 |
| EDN1 | FAXC | 0.648781 | 3 | 0 | 3 |
| EDN1 | NR2C2AP | 0.642801 | 3 | 0 | 3 |
| EDN1 | HCFC1 | 0.642229 | 3 | 0 | 3 |
For details and further investigation, click here