| Full name: mediator complex subunit 27 | Alias Symbol: TRAP37|CRSP34|MED3 | ||
| Type: protein-coding gene | Cytoband: 9q34.13 | ||
| Entrez ID: 9442 | HGNC ID: HGNC:2377 | Ensembl Gene: ENSG00000160563 | OMIM ID: 605044 |
| Drug and gene relationship at DGIdb | |||
Expression of MED27:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MED27 | 9442 | 51176_at | 0.1977 | 0.5940 | |
| GSE20347 | MED27 | 9442 | 51176_at | 0.2632 | 0.0743 | |
| GSE23400 | MED27 | 9442 | 51176_at | 0.0979 | 0.0029 | |
| GSE26886 | MED27 | 9442 | 51176_at | 0.7600 | 0.0002 | |
| GSE29001 | MED27 | 9442 | 51176_at | 0.2664 | 0.0541 | |
| GSE38129 | MED27 | 9442 | 51176_at | 0.4110 | 0.0018 | |
| GSE45670 | MED27 | 9442 | 51176_at | 0.3239 | 0.0507 | |
| GSE53622 | MED27 | 9442 | 67388 | 0.5982 | 0.0000 | |
| GSE53624 | MED27 | 9442 | 67388 | 0.5659 | 0.0000 | |
| GSE63941 | MED27 | 9442 | 51176_at | 0.7888 | 0.0215 | |
| GSE77861 | MED27 | 9442 | 51176_at | 0.2936 | 0.1379 | |
| GSE97050 | MED27 | 9442 | A_23_P301877 | 0.5544 | 0.1189 | |
| SRP007169 | MED27 | 9442 | RNAseq | 0.8893 | 0.0322 | |
| SRP008496 | MED27 | 9442 | RNAseq | 1.0088 | 0.0001 | |
| SRP064894 | MED27 | 9442 | RNAseq | 0.7909 | 0.0009 | |
| SRP133303 | MED27 | 9442 | RNAseq | 0.5679 | 0.0010 | |
| SRP159526 | MED27 | 9442 | RNAseq | 0.8978 | 0.0006 | |
| SRP193095 | MED27 | 9442 | RNAseq | 0.4618 | 0.0010 | |
| SRP219564 | MED27 | 9442 | RNAseq | 0.2970 | 0.3685 | |
| TCGA | MED27 | 9442 | RNAseq | 0.2992 | 0.0000 |
Upregulated datasets: 1; Downregulated datasets: 0.
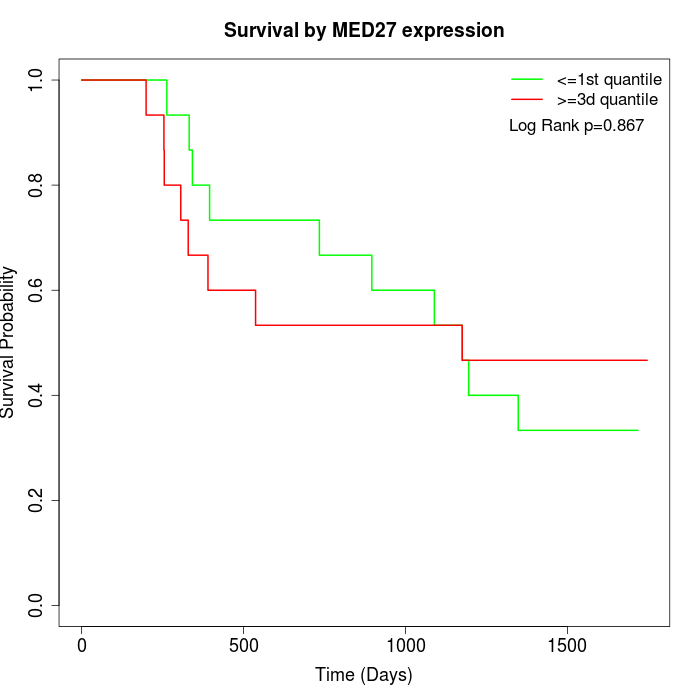
Survival by MED27 expression:
Note: Click image to view full size file.
Copy number change of MED27:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MED27 | 9442 | 7 | 8 | 15 | |
| GSE20123 | MED27 | 9442 | 7 | 8 | 15 | |
| GSE43470 | MED27 | 9442 | 5 | 7 | 31 | |
| GSE46452 | MED27 | 9442 | 6 | 13 | 40 | |
| GSE47630 | MED27 | 9442 | 4 | 15 | 21 | |
| GSE54993 | MED27 | 9442 | 3 | 3 | 64 | |
| GSE54994 | MED27 | 9442 | 12 | 9 | 32 | |
| GSE60625 | MED27 | 9442 | 0 | 0 | 11 | |
| GSE74703 | MED27 | 9442 | 5 | 5 | 26 | |
| GSE74704 | MED27 | 9442 | 4 | 6 | 10 | |
| TCGA | MED27 | 9442 | 28 | 26 | 42 |
Total number of gains: 81; Total number of losses: 100; Total Number of normals: 307.
Somatic mutations of MED27:
Generating mutation plots.
Highly correlated genes for MED27:
Showing top 20/1142 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MED27 | WIPI1 | 0.829624 | 3 | 0 | 3 |
| MED27 | PRELID1 | 0.805624 | 3 | 0 | 3 |
| MED27 | ZNF319 | 0.786786 | 3 | 0 | 3 |
| MED27 | CATSPER1 | 0.775125 | 3 | 0 | 3 |
| MED27 | C3orf33 | 0.750183 | 3 | 0 | 3 |
| MED27 | PIGS | 0.745313 | 3 | 0 | 3 |
| MED27 | SEMA3C | 0.738768 | 3 | 0 | 3 |
| MED27 | SLC39A3 | 0.734053 | 3 | 0 | 3 |
| MED27 | ACVRL1 | 0.729396 | 3 | 0 | 3 |
| MED27 | PLEKHM3 | 0.725039 | 3 | 0 | 3 |
| MED27 | ATXN3L | 0.715059 | 3 | 0 | 3 |
| MED27 | ELFN1 | 0.712035 | 3 | 0 | 3 |
| MED27 | TMEM38B | 0.708027 | 6 | 0 | 6 |
| MED27 | POLR2A | 0.703809 | 4 | 0 | 3 |
| MED27 | QSOX2 | 0.697628 | 5 | 0 | 4 |
| MED27 | NTMT1 | 0.693743 | 6 | 0 | 6 |
| MED27 | GNG10 | 0.692463 | 4 | 0 | 4 |
| MED27 | FBXW9 | 0.691893 | 3 | 0 | 3 |
| MED27 | ACAP2 | 0.691645 | 3 | 0 | 3 |
| MED27 | VBP1 | 0.691126 | 6 | 0 | 6 |
For details and further investigation, click here