| Full name: egl-9 family hypoxia inducible factor 1 | Alias Symbol: SM-20|PHD2|ZMYND6|HIFPH2 | ||
| Type: protein-coding gene | Cytoband: 1q42.2 | ||
| Entrez ID: 54583 | HGNC ID: HGNC:1232 | Ensembl Gene: ENSG00000135766 | OMIM ID: 606425 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
EGLN1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04066 | HIF-1 signaling pathway | |
| hsa05200 | Pathways in cancer | |
| hsa05211 | Renal cell carcinoma |
Expression of EGLN1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | EGLN1 | 54583 | 223046_at | -0.3464 | 0.4522 | |
| GSE20347 | EGLN1 | 54583 | 221497_x_at | 0.0905 | 0.6716 | |
| GSE23400 | EGLN1 | 54583 | 221497_x_at | 0.0470 | 0.5043 | |
| GSE26886 | EGLN1 | 54583 | 223046_at | -1.1751 | 0.0001 | |
| GSE29001 | EGLN1 | 54583 | 221497_x_at | 0.0426 | 0.9328 | |
| GSE38129 | EGLN1 | 54583 | 221497_x_at | 0.2249 | 0.1743 | |
| GSE45670 | EGLN1 | 54583 | 223046_at | -0.4537 | 0.0021 | |
| GSE53622 | EGLN1 | 54583 | 52233 | -0.1696 | 0.0012 | |
| GSE53624 | EGLN1 | 54583 | 52233 | -0.1514 | 0.0188 | |
| GSE63941 | EGLN1 | 54583 | 223046_at | -0.9493 | 0.0345 | |
| GSE77861 | EGLN1 | 54583 | 223046_at | -0.4101 | 0.2088 | |
| GSE97050 | EGLN1 | 54583 | A_23_P343935 | -0.4072 | 0.1453 | |
| SRP007169 | EGLN1 | 54583 | RNAseq | -1.0273 | 0.0043 | |
| SRP008496 | EGLN1 | 54583 | RNAseq | -0.9218 | 0.0001 | |
| SRP064894 | EGLN1 | 54583 | RNAseq | -0.3995 | 0.0306 | |
| SRP133303 | EGLN1 | 54583 | RNAseq | 0.0048 | 0.9678 | |
| SRP159526 | EGLN1 | 54583 | RNAseq | 0.0487 | 0.8858 | |
| SRP193095 | EGLN1 | 54583 | RNAseq | -0.4230 | 0.0002 | |
| SRP219564 | EGLN1 | 54583 | RNAseq | -0.2983 | 0.4631 | |
| TCGA | EGLN1 | 54583 | RNAseq | -0.1114 | 0.0164 |
Upregulated datasets: 0; Downregulated datasets: 2.
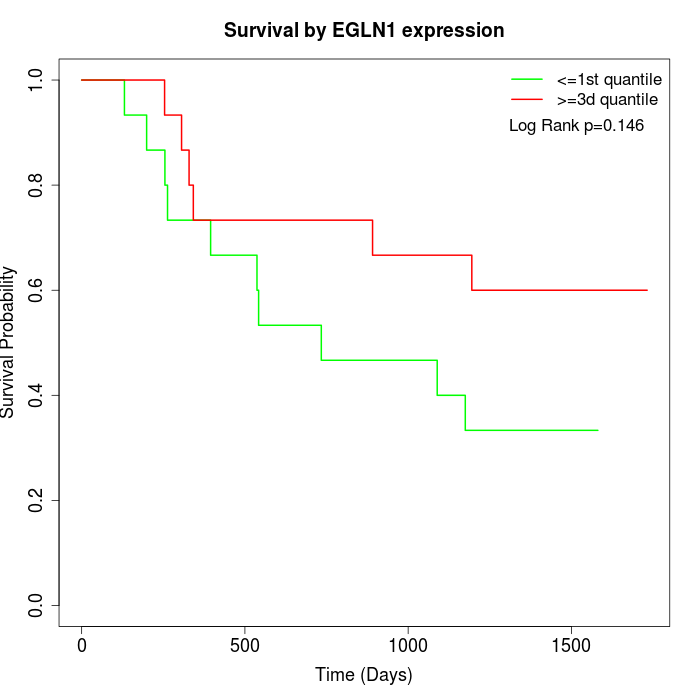
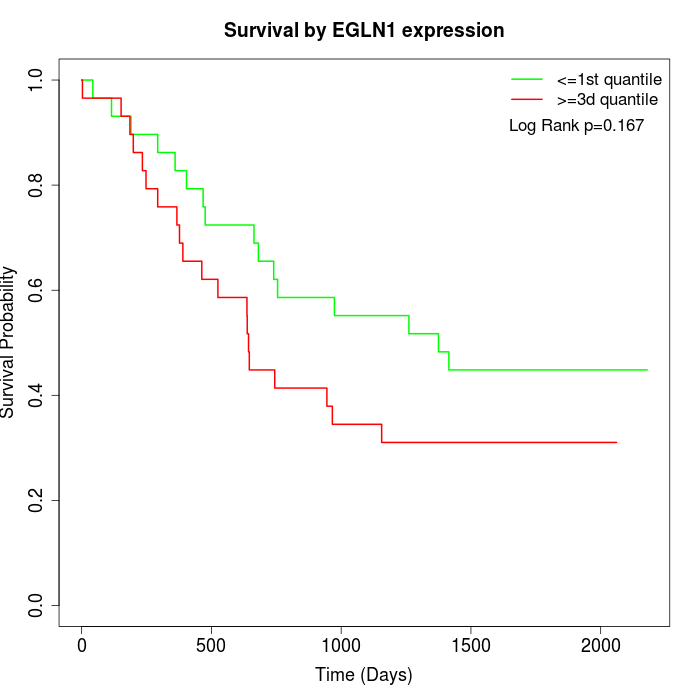
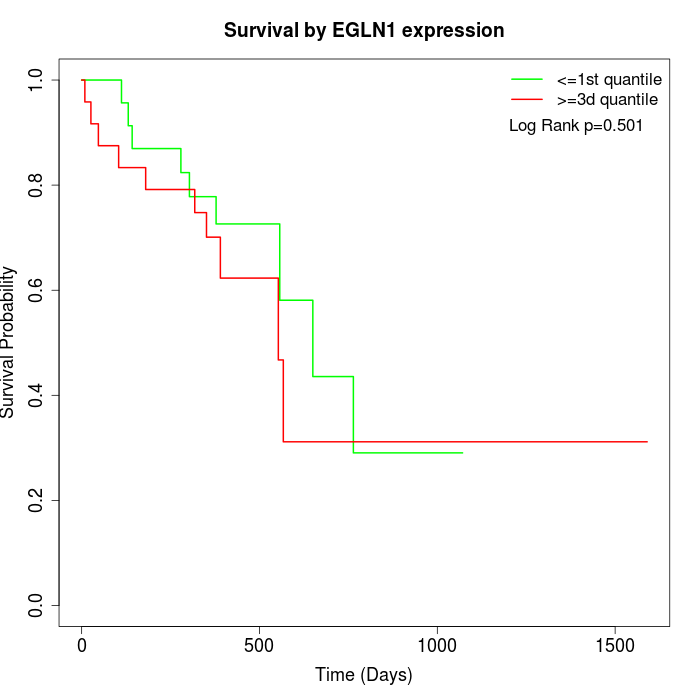
Survival by EGLN1 expression:
Note: Click image to view full size file.
Copy number change of EGLN1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | EGLN1 | 54583 | 12 | 0 | 18 | |
| GSE20123 | EGLN1 | 54583 | 12 | 0 | 18 | |
| GSE43470 | EGLN1 | 54583 | 8 | 1 | 34 | |
| GSE46452 | EGLN1 | 54583 | 4 | 2 | 53 | |
| GSE47630 | EGLN1 | 54583 | 15 | 0 | 25 | |
| GSE54993 | EGLN1 | 54583 | 0 | 6 | 64 | |
| GSE54994 | EGLN1 | 54583 | 17 | 0 | 36 | |
| GSE60625 | EGLN1 | 54583 | 0 | 0 | 11 | |
| GSE74703 | EGLN1 | 54583 | 8 | 1 | 27 | |
| GSE74704 | EGLN1 | 54583 | 6 | 0 | 14 | |
| TCGA | EGLN1 | 54583 | 45 | 3 | 48 |
Total number of gains: 127; Total number of losses: 13; Total Number of normals: 348.
Somatic mutations of EGLN1:
Generating mutation plots.
Highly correlated genes for EGLN1:
Showing top 20/547 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| EGLN1 | NBPF1 | 0.832087 | 3 | 0 | 3 |
| EGLN1 | ZNF879 | 0.799904 | 3 | 0 | 3 |
| EGLN1 | C2CD2 | 0.799424 | 3 | 0 | 3 |
| EGLN1 | RCHY1 | 0.796938 | 3 | 0 | 3 |
| EGLN1 | MFSD5 | 0.783368 | 3 | 0 | 3 |
| EGLN1 | ARFIP1 | 0.767103 | 3 | 0 | 3 |
| EGLN1 | SLC35B3 | 0.76453 | 3 | 0 | 3 |
| EGLN1 | RTN3 | 0.763747 | 3 | 0 | 3 |
| EGLN1 | SIK2 | 0.763329 | 3 | 0 | 3 |
| EGLN1 | TBC1D22B | 0.760426 | 3 | 0 | 3 |
| EGLN1 | TMEM50A | 0.7575 | 3 | 0 | 3 |
| EGLN1 | SLC25A27 | 0.753602 | 3 | 0 | 3 |
| EGLN1 | TAS2R5 | 0.738561 | 3 | 0 | 3 |
| EGLN1 | RCOR1 | 0.738209 | 4 | 0 | 3 |
| EGLN1 | CCNH | 0.73789 | 3 | 0 | 3 |
| EGLN1 | DOCK8 | 0.737223 | 3 | 0 | 3 |
| EGLN1 | NIPAL3 | 0.722132 | 3 | 0 | 3 |
| EGLN1 | DNAJB6 | 0.719774 | 4 | 0 | 3 |
| EGLN1 | GOLGA7 | 0.718 | 3 | 0 | 3 |
| EGLN1 | TBC1D20 | 0.717379 | 3 | 0 | 3 |
For details and further investigation, click here