| Full name: eukaryotic translation initiation factor 4B | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 12q13.13 | ||
| Entrez ID: 1975 | HGNC ID: HGNC:3285 | Ensembl Gene: ENSG00000063046 | OMIM ID: 603928 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
EIF4B involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04150 | mTOR signaling pathway | |
| hsa04151 | PI3K-Akt signaling pathway | |
| hsa05205 | Proteoglycans in cancer |
Expression of EIF4B:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | EIF4B | 1975 | 211938_at | -0.3714 | 0.2672 | |
| GSE20347 | EIF4B | 1975 | 211938_at | -0.4439 | 0.0002 | |
| GSE23400 | EIF4B | 1975 | 211938_at | -0.0858 | 0.2171 | |
| GSE26886 | EIF4B | 1975 | 211937_at | 0.3672 | 0.0657 | |
| GSE29001 | EIF4B | 1975 | 211938_at | -0.3735 | 0.1614 | |
| GSE38129 | EIF4B | 1975 | 211938_at | -0.3978 | 0.0002 | |
| GSE45670 | EIF4B | 1975 | 211938_at | -0.4421 | 0.0002 | |
| GSE53622 | EIF4B | 1975 | 117090 | -0.6150 | 0.0000 | |
| GSE53624 | EIF4B | 1975 | 117090 | -0.4597 | 0.0000 | |
| GSE63941 | EIF4B | 1975 | 211937_at | 1.1654 | 0.0160 | |
| GSE77861 | EIF4B | 1975 | 211938_at | -0.0630 | 0.7849 | |
| GSE97050 | EIF4B | 1975 | A_33_P3294133 | -0.5986 | 0.0976 | |
| SRP007169 | EIF4B | 1975 | RNAseq | -0.0126 | 0.9664 | |
| SRP008496 | EIF4B | 1975 | RNAseq | 0.0076 | 0.9682 | |
| SRP064894 | EIF4B | 1975 | RNAseq | -0.5978 | 0.0000 | |
| SRP133303 | EIF4B | 1975 | RNAseq | -0.5073 | 0.0000 | |
| SRP159526 | EIF4B | 1975 | RNAseq | -0.6481 | 0.0272 | |
| SRP193095 | EIF4B | 1975 | RNAseq | -0.6922 | 0.0000 | |
| SRP219564 | EIF4B | 1975 | RNAseq | -0.4582 | 0.0548 | |
| TCGA | EIF4B | 1975 | RNAseq | -0.1639 | 0.0000 |
Upregulated datasets: 1; Downregulated datasets: 0.
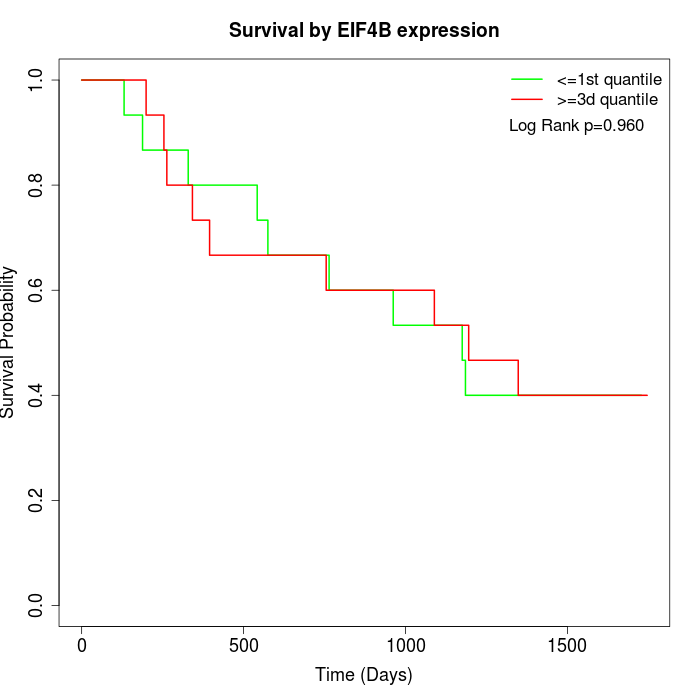
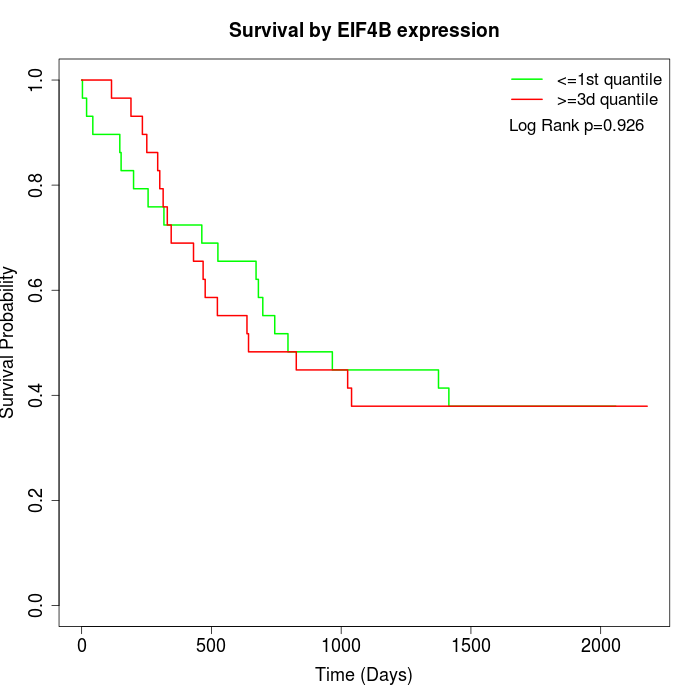
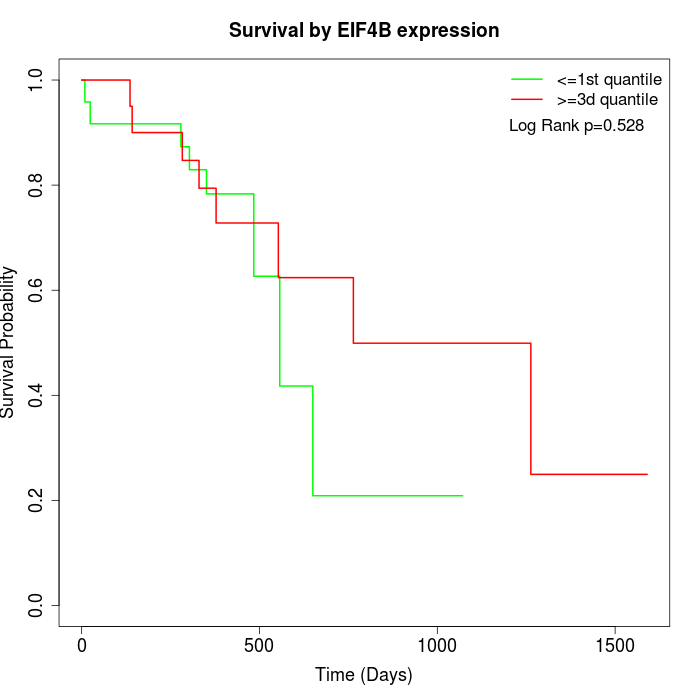
Survival by EIF4B expression:
Note: Click image to view full size file.
Copy number change of EIF4B:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | EIF4B | 1975 | 8 | 1 | 21 | |
| GSE20123 | EIF4B | 1975 | 8 | 1 | 21 | |
| GSE43470 | EIF4B | 1975 | 3 | 0 | 40 | |
| GSE46452 | EIF4B | 1975 | 7 | 1 | 51 | |
| GSE47630 | EIF4B | 1975 | 10 | 2 | 28 | |
| GSE54993 | EIF4B | 1975 | 0 | 5 | 65 | |
| GSE54994 | EIF4B | 1975 | 4 | 1 | 48 | |
| GSE60625 | EIF4B | 1975 | 0 | 0 | 11 | |
| GSE74703 | EIF4B | 1975 | 3 | 0 | 33 | |
| GSE74704 | EIF4B | 1975 | 5 | 1 | 14 | |
| TCGA | EIF4B | 1975 | 13 | 11 | 72 |
Total number of gains: 61; Total number of losses: 23; Total Number of normals: 404.
Somatic mutations of EIF4B:
Generating mutation plots.
Highly correlated genes for EIF4B:
Showing top 20/471 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| EIF4B | ITGA9 | 0.725013 | 4 | 0 | 4 |
| EIF4B | PPP1R12B | 0.707628 | 5 | 0 | 5 |
| EIF4B | RPL3 | 0.702394 | 9 | 0 | 8 |
| EIF4B | SMTN | 0.699203 | 3 | 0 | 3 |
| EIF4B | SORCS1 | 0.693588 | 3 | 0 | 3 |
| EIF4B | XPA | 0.690941 | 3 | 0 | 3 |
| EIF4B | CHRDL1 | 0.689019 | 4 | 0 | 4 |
| EIF4B | UBXN10 | 0.687137 | 3 | 0 | 3 |
| EIF4B | PREX2 | 0.686727 | 3 | 0 | 3 |
| EIF4B | DAZAP2 | 0.6851 | 3 | 0 | 3 |
| EIF4B | TXNIP | 0.683827 | 4 | 0 | 3 |
| EIF4B | RPS13 | 0.677587 | 3 | 0 | 3 |
| EIF4B | ZNF546 | 0.676736 | 3 | 0 | 3 |
| EIF4B | ARL5A | 0.673057 | 3 | 0 | 3 |
| EIF4B | ALDH1A2 | 0.671067 | 3 | 0 | 3 |
| EIF4B | VIT | 0.670382 | 4 | 0 | 4 |
| EIF4B | PWAR5 | 0.670168 | 3 | 0 | 3 |
| EIF4B | HSPB6 | 0.66477 | 4 | 0 | 3 |
| EIF4B | CGNL1 | 0.664541 | 4 | 0 | 3 |
| EIF4B | BEGAIN | 0.663205 | 3 | 0 | 3 |
For details and further investigation, click here