| Full name: integrin subunit alpha 9 | Alias Symbol: RLC|ITGA4L|ALPHA-RLC | ||
| Type: protein-coding gene | Cytoband: 3p22.2 | ||
| Entrez ID: 3680 | HGNC ID: HGNC:6145 | Ensembl Gene: ENSG00000144668 | OMIM ID: 603963 |
| Drug and gene relationship at DGIdb | |||
ITGA9 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04151 | PI3K-Akt signaling pathway | |
| hsa04510 | Focal adhesion | |
| hsa04810 | Regulation of actin cytoskeleton |
Expression of ITGA9:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ITGA9 | 3680 | 227297_at | -1.7289 | 0.0962 | |
| GSE20347 | ITGA9 | 3680 | 206009_at | -0.0403 | 0.7642 | |
| GSE23400 | ITGA9 | 3680 | 206009_at | -0.1874 | 0.0000 | |
| GSE26886 | ITGA9 | 3680 | 227297_at | 0.3137 | 0.4270 | |
| GSE29001 | ITGA9 | 3680 | 206009_at | -0.0512 | 0.8226 | |
| GSE38129 | ITGA9 | 3680 | 206009_at | -0.3257 | 0.0046 | |
| GSE45670 | ITGA9 | 3680 | 227297_at | -2.6472 | 0.0000 | |
| GSE53622 | ITGA9 | 3680 | 60819 | -1.6088 | 0.0000 | |
| GSE53624 | ITGA9 | 3680 | 60819 | -1.1195 | 0.0000 | |
| GSE63941 | ITGA9 | 3680 | 206009_at | 0.0121 | 0.9634 | |
| GSE77861 | ITGA9 | 3680 | 206009_at | -0.0972 | 0.6005 | |
| GSE97050 | ITGA9 | 3680 | A_23_P252193 | -1.0574 | 0.0844 | |
| SRP007169 | ITGA9 | 3680 | RNAseq | 1.2508 | 0.0131 | |
| SRP008496 | ITGA9 | 3680 | RNAseq | 1.9389 | 0.0001 | |
| SRP064894 | ITGA9 | 3680 | RNAseq | -1.1216 | 0.0012 | |
| SRP133303 | ITGA9 | 3680 | RNAseq | -0.9795 | 0.0374 | |
| SRP159526 | ITGA9 | 3680 | RNAseq | -0.0600 | 0.9320 | |
| SRP193095 | ITGA9 | 3680 | RNAseq | -0.0366 | 0.8767 | |
| SRP219564 | ITGA9 | 3680 | RNAseq | -1.0980 | 0.1970 | |
| TCGA | ITGA9 | 3680 | RNAseq | -1.0276 | 0.0001 |
Upregulated datasets: 2; Downregulated datasets: 5.
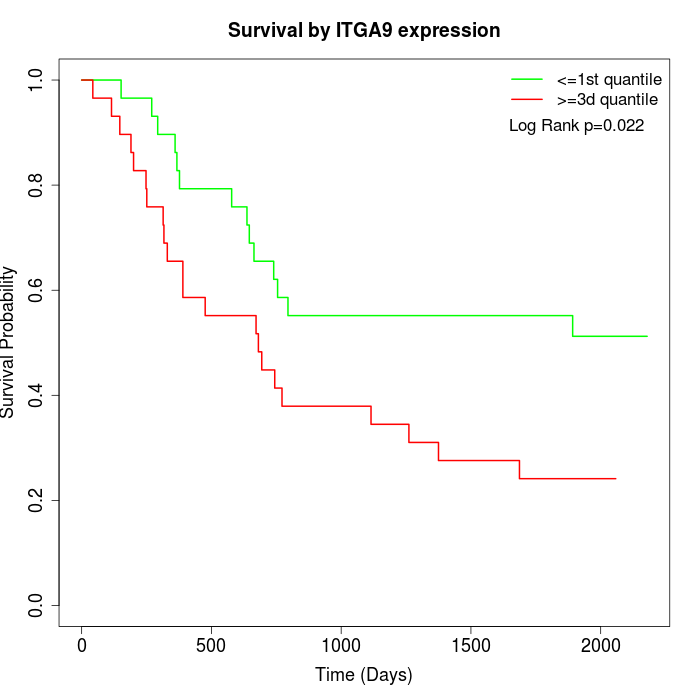
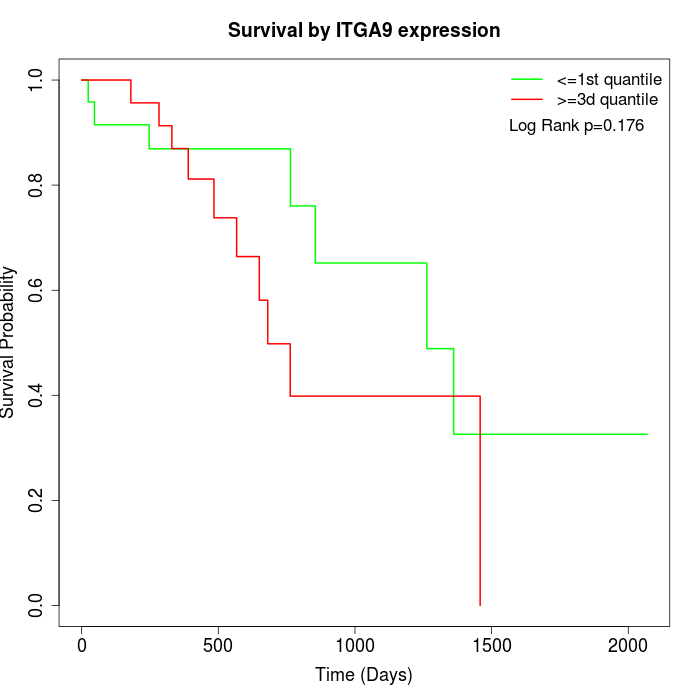
Survival by ITGA9 expression:
Note: Click image to view full size file.
Copy number change of ITGA9:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ITGA9 | 3680 | 0 | 19 | 11 | |
| GSE20123 | ITGA9 | 3680 | 0 | 19 | 11 | |
| GSE43470 | ITGA9 | 3680 | 0 | 20 | 23 | |
| GSE46452 | ITGA9 | 3680 | 2 | 17 | 40 | |
| GSE47630 | ITGA9 | 3680 | 1 | 25 | 14 | |
| GSE54993 | ITGA9 | 3680 | 6 | 3 | 61 | |
| GSE54994 | ITGA9 | 3680 | 1 | 32 | 20 | |
| GSE60625 | ITGA9 | 3680 | 5 | 0 | 6 | |
| GSE74703 | ITGA9 | 3680 | 0 | 16 | 20 | |
| GSE74704 | ITGA9 | 3680 | 0 | 13 | 7 | |
| TCGA | ITGA9 | 3680 | 0 | 72 | 24 |
Total number of gains: 15; Total number of losses: 236; Total Number of normals: 237.
Somatic mutations of ITGA9:
Generating mutation plots.
Highly correlated genes for ITGA9:
Showing top 20/1222 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ITGA9 | MSRB3 | 0.854613 | 5 | 0 | 5 |
| ITGA9 | MAMDC2 | 0.852152 | 5 | 0 | 5 |
| ITGA9 | SYNPO2 | 0.845464 | 6 | 0 | 6 |
| ITGA9 | NEGR1 | 0.83268 | 5 | 0 | 5 |
| ITGA9 | NEXN | 0.831365 | 6 | 0 | 6 |
| ITGA9 | UBXN10 | 0.829079 | 3 | 0 | 3 |
| ITGA9 | C1QTNF7 | 0.828933 | 5 | 0 | 5 |
| ITGA9 | MRVI1 | 0.822495 | 6 | 0 | 5 |
| ITGA9 | MACROD2 | 0.820651 | 3 | 0 | 3 |
| ITGA9 | TIMP3 | 0.81308 | 3 | 0 | 3 |
| ITGA9 | CACNB2 | 0.805349 | 6 | 0 | 6 |
| ITGA9 | EIF4E3 | 0.801119 | 5 | 0 | 5 |
| ITGA9 | CYP4V2 | 0.799469 | 3 | 0 | 3 |
| ITGA9 | RNF150 | 0.796451 | 6 | 0 | 6 |
| ITGA9 | LRFN5 | 0.794976 | 4 | 0 | 4 |
| ITGA9 | CLEC3B | 0.791719 | 3 | 0 | 3 |
| ITGA9 | TNFSF12 | 0.791277 | 3 | 0 | 3 |
| ITGA9 | MAGI2-AS3 | 0.786319 | 4 | 0 | 4 |
| ITGA9 | FBXL3 | 0.782467 | 3 | 0 | 3 |
| ITGA9 | FHL1 | 0.779562 | 7 | 0 | 6 |
For details and further investigation, click here