| Full name: ERGIC and golgi 2 | Alias Symbol: PTX1|Erv41 | ||
| Type: protein-coding gene | Cytoband: 12p11.22 | ||
| Entrez ID: 51290 | HGNC ID: HGNC:30208 | Ensembl Gene: ENSG00000087502 | OMIM ID: 612236 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of ERGIC2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ERGIC2 | 51290 | 226422_at | 0.1638 | 0.4665 | |
| GSE20347 | ERGIC2 | 51290 | 218135_at | 0.1180 | 0.6838 | |
| GSE23400 | ERGIC2 | 51290 | 218135_at | 0.2344 | 0.0672 | |
| GSE26886 | ERGIC2 | 51290 | 226422_at | 0.3661 | 0.1036 | |
| GSE29001 | ERGIC2 | 51290 | 218135_at | 0.4451 | 0.2907 | |
| GSE38129 | ERGIC2 | 51290 | 218135_at | 0.2064 | 0.4427 | |
| GSE45670 | ERGIC2 | 51290 | 218135_at | 0.1855 | 0.2914 | |
| GSE53622 | ERGIC2 | 51290 | 15286 | 0.3915 | 0.0000 | |
| GSE53624 | ERGIC2 | 51290 | 15286 | 0.3202 | 0.0000 | |
| GSE63941 | ERGIC2 | 51290 | 226422_at | -0.2204 | 0.6940 | |
| GSE77861 | ERGIC2 | 51290 | 226422_at | 0.3205 | 0.0796 | |
| GSE97050 | ERGIC2 | 51290 | A_23_P139509 | 0.2383 | 0.3004 | |
| SRP007169 | ERGIC2 | 51290 | RNAseq | 0.0462 | 0.8889 | |
| SRP008496 | ERGIC2 | 51290 | RNAseq | 0.3148 | 0.1798 | |
| SRP064894 | ERGIC2 | 51290 | RNAseq | 0.1355 | 0.3254 | |
| SRP133303 | ERGIC2 | 51290 | RNAseq | 0.3960 | 0.0158 | |
| SRP159526 | ERGIC2 | 51290 | RNAseq | 0.2174 | 0.1931 | |
| SRP193095 | ERGIC2 | 51290 | RNAseq | -0.2400 | 0.0858 | |
| SRP219564 | ERGIC2 | 51290 | RNAseq | -0.1762 | 0.5885 | |
| TCGA | ERGIC2 | 51290 | RNAseq | 0.0970 | 0.0665 |
Upregulated datasets: 0; Downregulated datasets: 0.
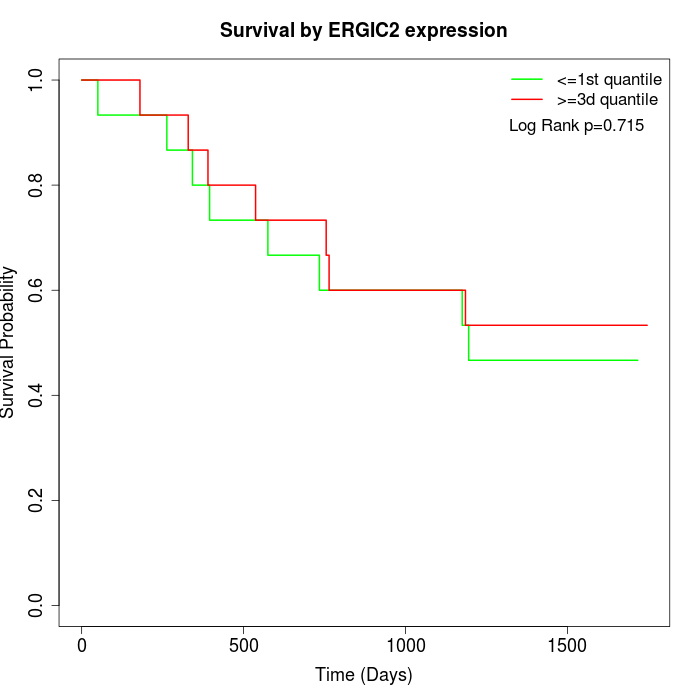
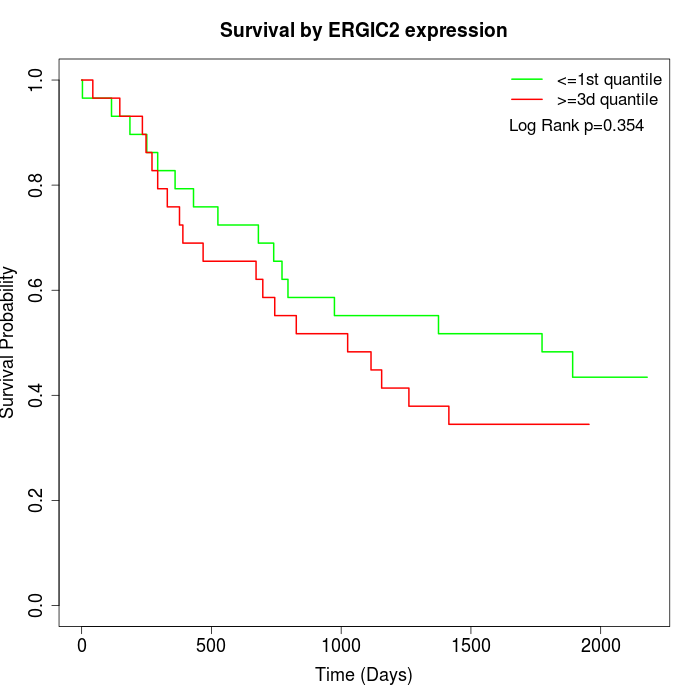
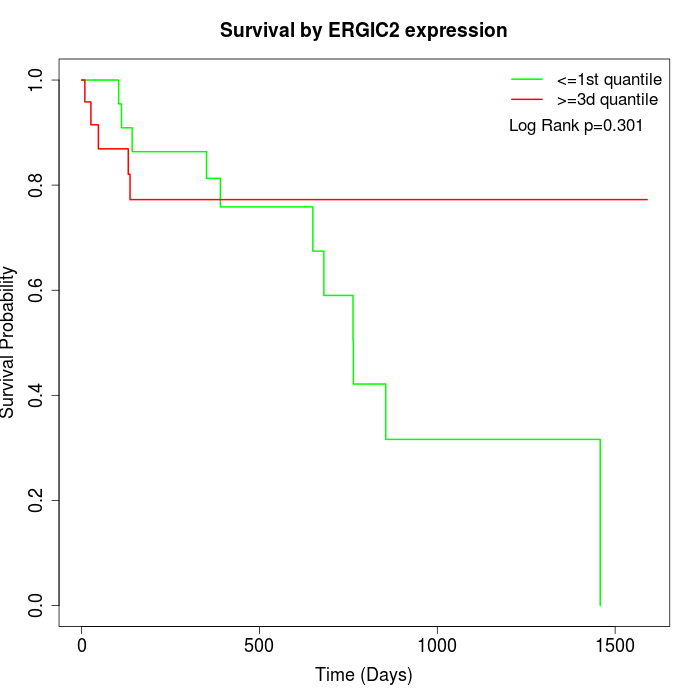
Survival by ERGIC2 expression:
Note: Click image to view full size file.
Copy number change of ERGIC2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ERGIC2 | 51290 | 9 | 1 | 20 | |
| GSE20123 | ERGIC2 | 51290 | 9 | 1 | 20 | |
| GSE43470 | ERGIC2 | 51290 | 8 | 3 | 32 | |
| GSE46452 | ERGIC2 | 51290 | 10 | 1 | 48 | |
| GSE47630 | ERGIC2 | 51290 | 13 | 0 | 27 | |
| GSE54993 | ERGIC2 | 51290 | 0 | 9 | 61 | |
| GSE54994 | ERGIC2 | 51290 | 9 | 2 | 42 | |
| GSE60625 | ERGIC2 | 51290 | 0 | 1 | 10 | |
| GSE74703 | ERGIC2 | 51290 | 7 | 2 | 27 | |
| GSE74704 | ERGIC2 | 51290 | 5 | 1 | 14 | |
| TCGA | ERGIC2 | 51290 | 41 | 7 | 48 |
Total number of gains: 111; Total number of losses: 28; Total Number of normals: 349.
Somatic mutations of ERGIC2:
Generating mutation plots.
Highly correlated genes for ERGIC2:
Showing top 20/866 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ERGIC2 | TPRA1 | 0.806862 | 3 | 0 | 3 |
| ERGIC2 | RNF138 | 0.801156 | 3 | 0 | 3 |
| ERGIC2 | ZNF124 | 0.80058 | 3 | 0 | 3 |
| ERGIC2 | SIPA1L2 | 0.796394 | 3 | 0 | 3 |
| ERGIC2 | U2AF1 | 0.788646 | 3 | 0 | 3 |
| ERGIC2 | ZNF567 | 0.785368 | 3 | 0 | 3 |
| ERGIC2 | FLYWCH2 | 0.76408 | 3 | 0 | 3 |
| ERGIC2 | C1orf35 | 0.759176 | 3 | 0 | 3 |
| ERGIC2 | GOLPH3 | 0.755217 | 3 | 0 | 3 |
| ERGIC2 | RNF4 | 0.749773 | 3 | 0 | 3 |
| ERGIC2 | ANKIB1 | 0.738818 | 3 | 0 | 3 |
| ERGIC2 | LSM4 | 0.733206 | 3 | 0 | 3 |
| ERGIC2 | ANKRD54 | 0.722097 | 3 | 0 | 3 |
| ERGIC2 | SP100 | 0.719402 | 3 | 0 | 3 |
| ERGIC2 | PDK1 | 0.717038 | 3 | 0 | 3 |
| ERGIC2 | AP1AR | 0.716352 | 3 | 0 | 3 |
| ERGIC2 | MCCC1 | 0.705448 | 3 | 0 | 3 |
| ERGIC2 | MED21 | 0.705343 | 3 | 0 | 3 |
| ERGIC2 | MRPL35 | 0.701997 | 4 | 0 | 4 |
| ERGIC2 | CHAMP1 | 0.70158 | 3 | 0 | 3 |
For details and further investigation, click here