| Full name: ezrin | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 6q25.3 | ||
| Entrez ID: 7430 | HGNC ID: HGNC:12691 | Ensembl Gene: ENSG00000092820 | OMIM ID: 123900 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
EZR involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04670 | Leukocyte transendothelial migration | |
| hsa04810 | Regulation of actin cytoskeleton | |
| hsa04971 | Gastric acid secretion | |
| hsa05205 | Proteoglycans in cancer |
Expression of EZR:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | EZR | 7430 | 208623_s_at | -0.6022 | 0.1017 | |
| GSE20347 | EZR | 7430 | 208623_s_at | -0.7986 | 0.0000 | |
| GSE23400 | EZR | 7430 | 208623_s_at | -0.5604 | 0.0000 | |
| GSE26886 | EZR | 7430 | 208623_s_at | -0.6623 | 0.0083 | |
| GSE29001 | EZR | 7430 | 208623_s_at | -1.1132 | 0.0234 | |
| GSE38129 | EZR | 7430 | 208623_s_at | -0.4983 | 0.0020 | |
| GSE45670 | EZR | 7430 | 208623_s_at | -0.0907 | 0.5988 | |
| GSE53622 | EZR | 7430 | 120165 | -0.8256 | 0.0000 | |
| GSE53624 | EZR | 7430 | 120165 | -0.8583 | 0.0000 | |
| GSE63941 | EZR | 7430 | 208623_s_at | 2.0368 | 0.0002 | |
| GSE77861 | EZR | 7430 | 208623_s_at | -0.3407 | 0.1944 | |
| GSE97050 | EZR | 7430 | A_23_P19590 | 0.0026 | 0.9955 | |
| SRP007169 | EZR | 7430 | RNAseq | -0.8121 | 0.0686 | |
| SRP008496 | EZR | 7430 | RNAseq | -0.8398 | 0.0007 | |
| SRP064894 | EZR | 7430 | RNAseq | -1.1524 | 0.0000 | |
| SRP133303 | EZR | 7430 | RNAseq | -0.5300 | 0.0046 | |
| SRP159526 | EZR | 7430 | RNAseq | -1.3923 | 0.0000 | |
| SRP193095 | EZR | 7430 | RNAseq | -0.8154 | 0.0000 | |
| SRP219564 | EZR | 7430 | RNAseq | -0.9698 | 0.0146 | |
| TCGA | EZR | 7430 | RNAseq | -0.2443 | 0.0000 |
Upregulated datasets: 1; Downregulated datasets: 3.
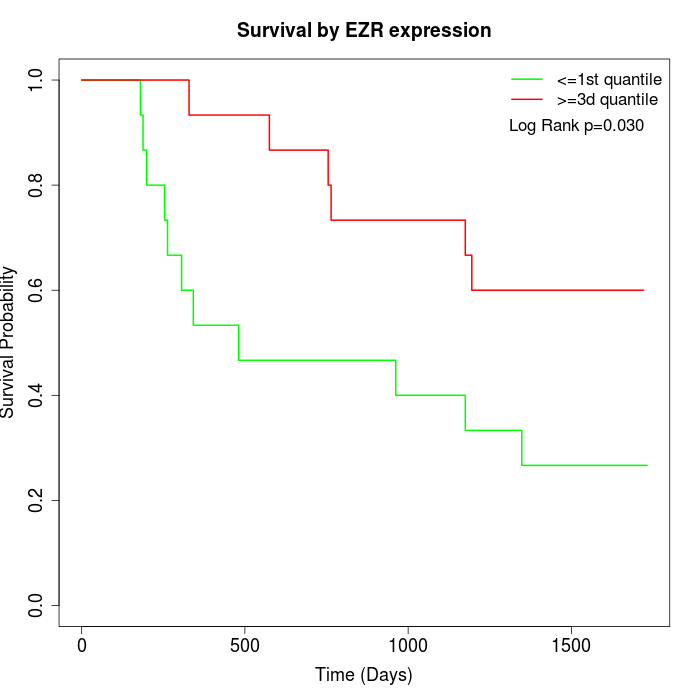
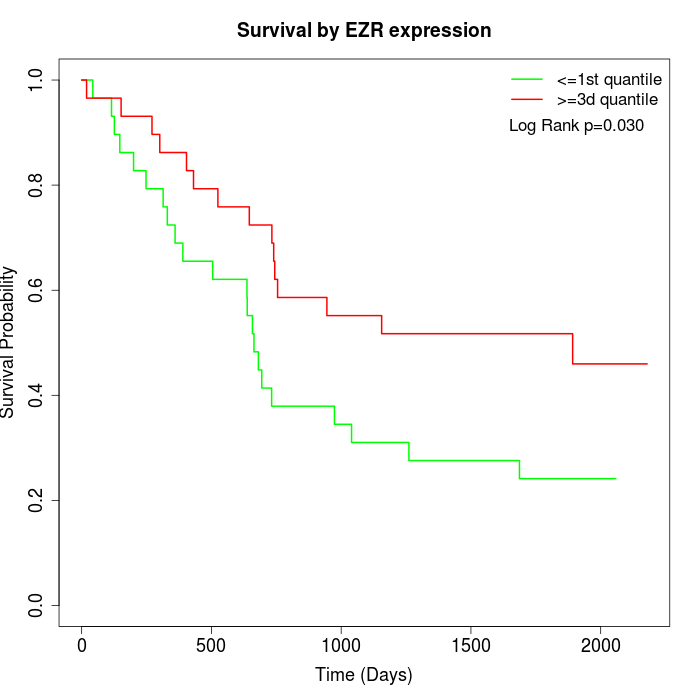
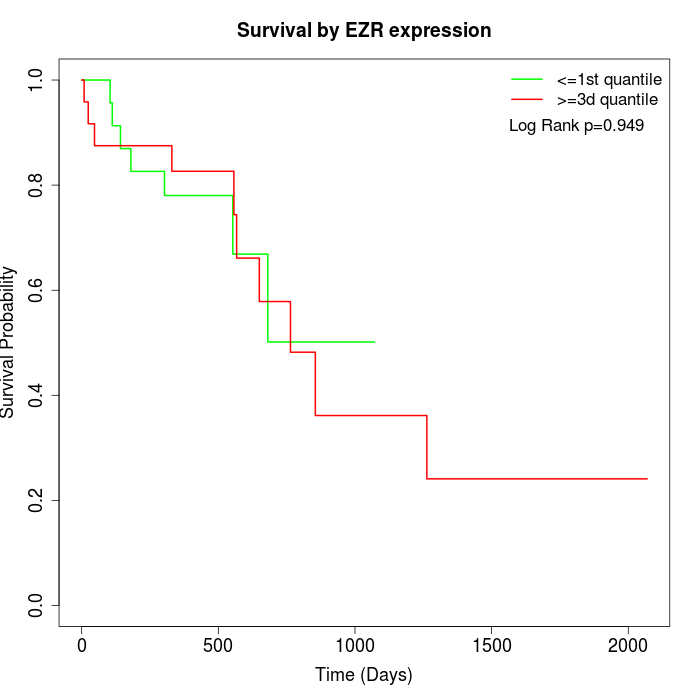
Survival by EZR expression:
Note: Click image to view full size file.
Copy number change of EZR:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | EZR | 7430 | 3 | 5 | 22 | |
| GSE20123 | EZR | 7430 | 3 | 4 | 23 | |
| GSE43470 | EZR | 7430 | 4 | 0 | 39 | |
| GSE46452 | EZR | 7430 | 3 | 10 | 46 | |
| GSE47630 | EZR | 7430 | 9 | 4 | 27 | |
| GSE54993 | EZR | 7430 | 3 | 2 | 65 | |
| GSE54994 | EZR | 7430 | 8 | 8 | 37 | |
| GSE60625 | EZR | 7430 | 0 | 1 | 10 | |
| GSE74703 | EZR | 7430 | 4 | 0 | 32 | |
| GSE74704 | EZR | 7430 | 1 | 2 | 17 | |
| TCGA | EZR | 7430 | 14 | 21 | 61 |
Total number of gains: 52; Total number of losses: 57; Total Number of normals: 379.
Somatic mutations of EZR:
Generating mutation plots.
Highly correlated genes for EZR:
Showing top 20/1215 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| EZR | NUAK2 | 0.763023 | 3 | 0 | 3 |
| EZR | UCHL5 | 0.762649 | 3 | 0 | 3 |
| EZR | COPS4 | 0.747949 | 3 | 0 | 3 |
| EZR | ELOVL1 | 0.747914 | 3 | 0 | 3 |
| EZR | PXMP2 | 0.74573 | 3 | 0 | 3 |
| EZR | CPOX | 0.741776 | 3 | 0 | 3 |
| EZR | KDM3A | 0.734562 | 3 | 0 | 3 |
| EZR | CHCHD1 | 0.734344 | 3 | 0 | 3 |
| EZR | YTHDF2 | 0.7273 | 3 | 0 | 3 |
| EZR | CA5B | 0.727004 | 3 | 0 | 3 |
| EZR | AIMP1 | 0.723995 | 3 | 0 | 3 |
| EZR | TMEM154 | 0.715312 | 6 | 0 | 6 |
| EZR | MCTS1 | 0.714261 | 3 | 0 | 3 |
| EZR | UBQLN1 | 0.713412 | 3 | 0 | 3 |
| EZR | NIPAL1 | 0.709338 | 3 | 0 | 3 |
| EZR | EPGN | 0.708205 | 3 | 0 | 3 |
| EZR | RAB11A | 0.700821 | 11 | 0 | 11 |
| EZR | BLNK | 0.699871 | 11 | 0 | 11 |
| EZR | STAMBP | 0.699396 | 3 | 0 | 3 |
| EZR | ZNF165 | 0.698261 | 3 | 0 | 3 |
For details and further investigation, click here