| Full name: peroxidasin | Alias Symbol: KIAA0230|PRG2|MG50|D2S448|D2S448E|PXN | ||
| Type: protein-coding gene | Cytoband: 2p25.3 | ||
| Entrez ID: 7837 | HGNC ID: HGNC:14966 | Ensembl Gene: ENSG00000130508 | OMIM ID: 605158 |
| Drug and gene relationship at DGIdb | |||
Expression of PXDN:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PXDN | 7837 | 212012_at | 0.7573 | 0.3520 | |
| GSE20347 | PXDN | 7837 | 212012_at | 1.7273 | 0.0000 | |
| GSE23400 | PXDN | 7837 | 212012_at | 1.3013 | 0.0000 | |
| GSE26886 | PXDN | 7837 | 212012_at | 3.5818 | 0.0000 | |
| GSE29001 | PXDN | 7837 | 212012_at | 2.3638 | 0.0001 | |
| GSE38129 | PXDN | 7837 | 212012_at | 1.6519 | 0.0001 | |
| GSE45670 | PXDN | 7837 | 212012_at | 1.2177 | 0.0159 | |
| GSE53622 | PXDN | 7837 | 58560 | 1.8865 | 0.0000 | |
| GSE53624 | PXDN | 7837 | 58560 | 2.3493 | 0.0000 | |
| GSE63941 | PXDN | 7837 | 212012_at | -4.5867 | 0.0103 | |
| GSE77861 | PXDN | 7837 | 212012_at | 1.2534 | 0.0069 | |
| GSE97050 | PXDN | 7837 | A_33_P3238166 | 1.1244 | 0.0620 | |
| SRP007169 | PXDN | 7837 | RNAseq | 7.0149 | 0.0000 | |
| SRP008496 | PXDN | 7837 | RNAseq | 5.9404 | 0.0000 | |
| SRP064894 | PXDN | 7837 | RNAseq | 2.4899 | 0.0000 | |
| SRP133303 | PXDN | 7837 | RNAseq | 2.1796 | 0.0000 | |
| SRP159526 | PXDN | 7837 | RNAseq | 3.1198 | 0.0000 | |
| SRP193095 | PXDN | 7837 | RNAseq | 3.1272 | 0.0000 | |
| SRP219564 | PXDN | 7837 | RNAseq | 1.0583 | 0.1102 | |
| TCGA | PXDN | 7837 | RNAseq | 0.3539 | 0.0034 |
Upregulated datasets: 15; Downregulated datasets: 1.
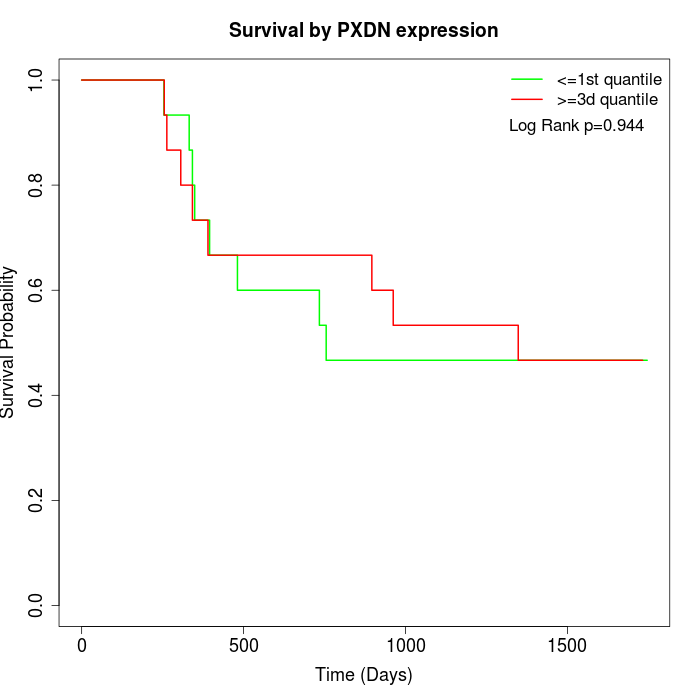
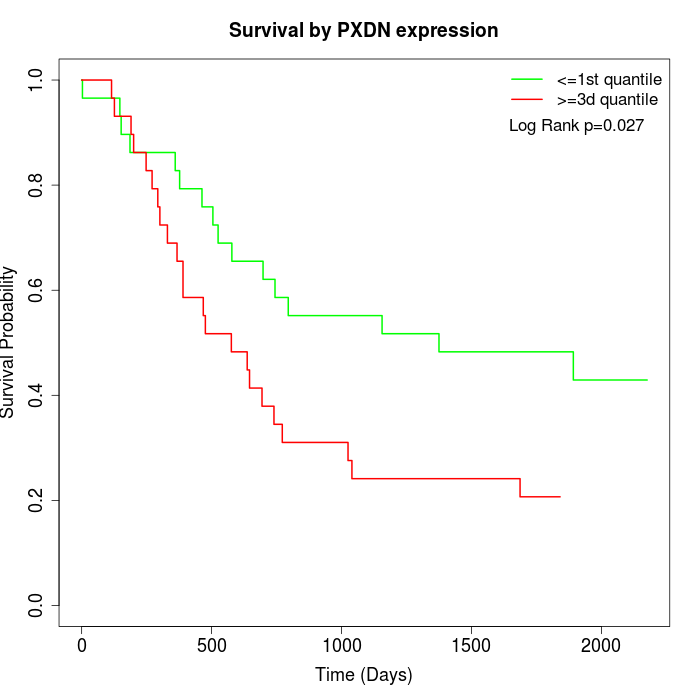
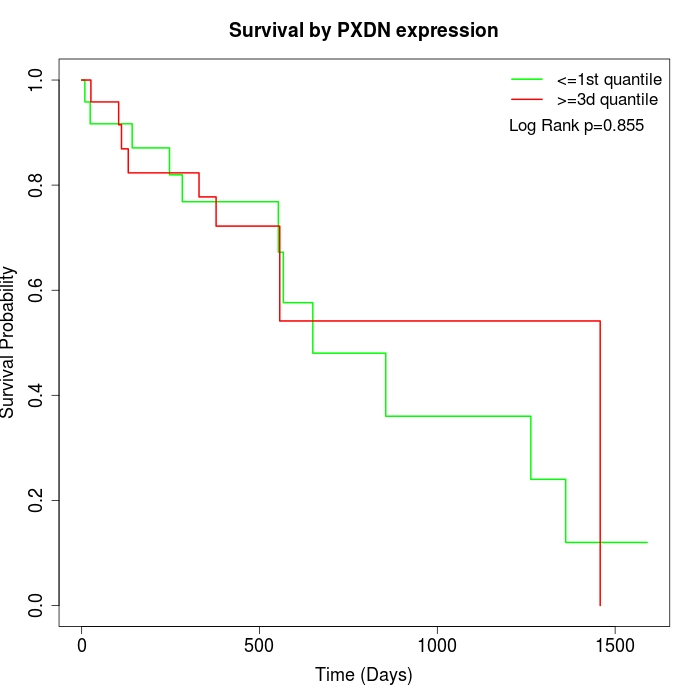
Survival by PXDN expression:
Note: Click image to view full size file.
Copy number change of PXDN:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PXDN | 7837 | 8 | 3 | 19 | |
| GSE20123 | PXDN | 7837 | 8 | 3 | 19 | |
| GSE43470 | PXDN | 7837 | 3 | 1 | 39 | |
| GSE46452 | PXDN | 7837 | 3 | 4 | 52 | |
| GSE47630 | PXDN | 7837 | 7 | 0 | 33 | |
| GSE54993 | PXDN | 7837 | 1 | 7 | 62 | |
| GSE54994 | PXDN | 7837 | 13 | 0 | 40 | |
| GSE60625 | PXDN | 7837 | 0 | 3 | 8 | |
| GSE74703 | PXDN | 7837 | 3 | 0 | 33 | |
| GSE74704 | PXDN | 7837 | 8 | 1 | 11 | |
| TCGA | PXDN | 7837 | 32 | 5 | 59 |
Total number of gains: 86; Total number of losses: 27; Total Number of normals: 375.
Somatic mutations of PXDN:
Generating mutation plots.
Highly correlated genes for PXDN:
Showing top 20/1667 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PXDN | WDR54 | 0.815851 | 7 | 0 | 7 |
| PXDN | VCAN | 0.81204 | 13 | 0 | 13 |
| PXDN | FADS1 | 0.805662 | 3 | 0 | 3 |
| PXDN | FCGR3A | 0.801522 | 3 | 0 | 3 |
| PXDN | COL4A1 | 0.801519 | 13 | 0 | 12 |
| PXDN | C11orf96 | 0.793814 | 3 | 0 | 3 |
| PXDN | TRAM2 | 0.792018 | 12 | 0 | 11 |
| PXDN | CDH11 | 0.786677 | 13 | 0 | 12 |
| PXDN | RAI14 | 0.781787 | 12 | 0 | 12 |
| PXDN | SLC39A14 | 0.780966 | 12 | 0 | 11 |
| PXDN | MMD | 0.780743 | 13 | 0 | 12 |
| PXDN | LOXL2 | 0.780705 | 12 | 0 | 12 |
| PXDN | CALU | 0.780205 | 12 | 0 | 12 |
| PXDN | GNG10 | 0.778051 | 3 | 0 | 3 |
| PXDN | SHISA2 | 0.772183 | 6 | 0 | 6 |
| PXDN | NUAK1 | 0.7706 | 12 | 0 | 11 |
| PXDN | LPCAT1 | 0.770059 | 12 | 0 | 12 |
| PXDN | PLOD3 | 0.766734 | 11 | 0 | 11 |
| PXDN | COL5A1 | 0.760965 | 13 | 0 | 13 |
| PXDN | MFAP2 | 0.75998 | 12 | 0 | 11 |
For details and further investigation, click here