| Full name: lysophosphatidylcholine acyltransferase 1 | Alias Symbol: FLJ12443|AGPAT9|AGPAT10 | ||
| Type: protein-coding gene | Cytoband: 5p15.33 | ||
| Entrez ID: 79888 | HGNC ID: HGNC:25718 | Ensembl Gene: ENSG00000153395 | OMIM ID: 610472 |
| Drug and gene relationship at DGIdb | |||
Expression of LPCAT1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LPCAT1 | 79888 | 201818_at | 1.6488 | 0.0420 | |
| GSE20347 | LPCAT1 | 79888 | 201818_at | 1.9929 | 0.0000 | |
| GSE23400 | LPCAT1 | 79888 | 201818_at | 1.0446 | 0.0000 | |
| GSE26886 | LPCAT1 | 79888 | 201818_at | 2.9095 | 0.0000 | |
| GSE29001 | LPCAT1 | 79888 | 201818_at | 1.7308 | 0.0000 | |
| GSE38129 | LPCAT1 | 79888 | 201818_at | 1.6244 | 0.0000 | |
| GSE45670 | LPCAT1 | 79888 | 201818_at | 1.4592 | 0.0001 | |
| GSE53622 | LPCAT1 | 79888 | 13931 | 1.7278 | 0.0000 | |
| GSE53624 | LPCAT1 | 79888 | 13931 | 2.0754 | 0.0000 | |
| GSE63941 | LPCAT1 | 79888 | 201818_at | -0.5231 | 0.4151 | |
| GSE77861 | LPCAT1 | 79888 | 201818_at | 1.6719 | 0.0008 | |
| GSE97050 | LPCAT1 | 79888 | A_24_P406006 | 1.2659 | 0.0620 | |
| SRP007169 | LPCAT1 | 79888 | RNAseq | 4.1114 | 0.0000 | |
| SRP008496 | LPCAT1 | 79888 | RNAseq | 3.8049 | 0.0000 | |
| SRP064894 | LPCAT1 | 79888 | RNAseq | 1.7656 | 0.0000 | |
| SRP133303 | LPCAT1 | 79888 | RNAseq | 1.7773 | 0.0000 | |
| SRP159526 | LPCAT1 | 79888 | RNAseq | 1.4327 | 0.0000 | |
| SRP193095 | LPCAT1 | 79888 | RNAseq | 2.0518 | 0.0000 | |
| SRP219564 | LPCAT1 | 79888 | RNAseq | 1.5828 | 0.0123 | |
| TCGA | LPCAT1 | 79888 | RNAseq | 0.3056 | 0.0003 |
Upregulated datasets: 17; Downregulated datasets: 0.
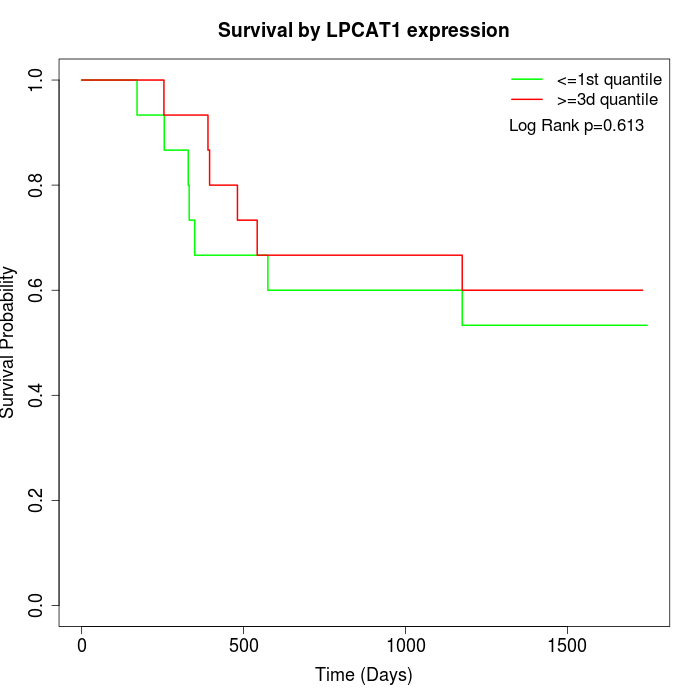
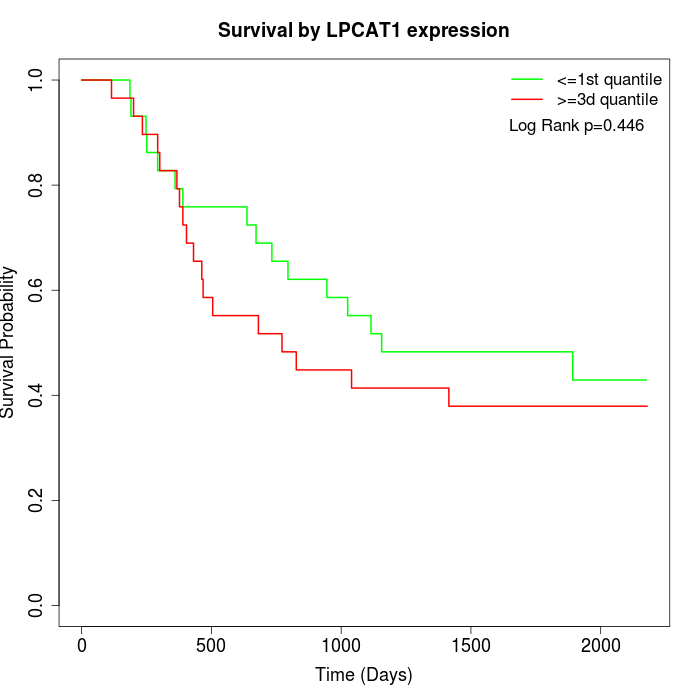
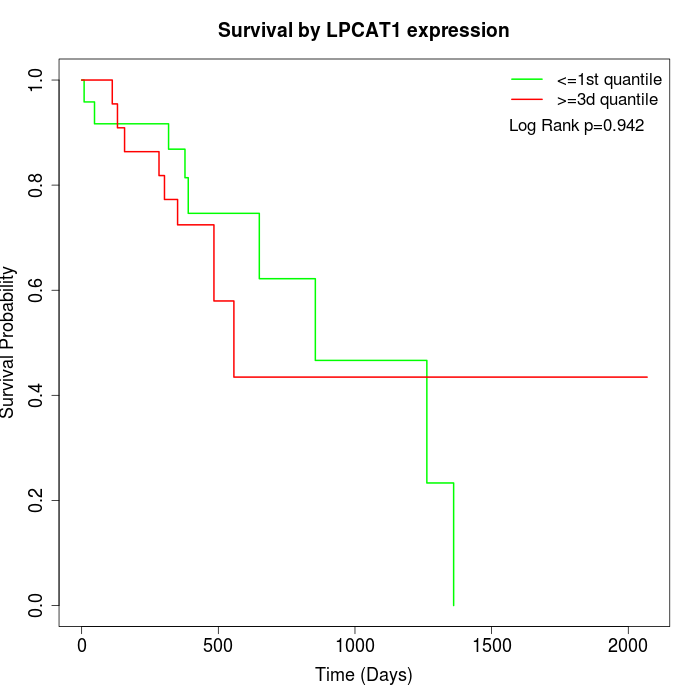
Survival by LPCAT1 expression:
Note: Click image to view full size file.
Copy number change of LPCAT1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LPCAT1 | 79888 | 13 | 1 | 16 | |
| GSE20123 | LPCAT1 | 79888 | 13 | 1 | 16 | |
| GSE43470 | LPCAT1 | 79888 | 17 | 1 | 25 | |
| GSE46452 | LPCAT1 | 79888 | 6 | 22 | 31 | |
| GSE47630 | LPCAT1 | 79888 | 7 | 11 | 22 | |
| GSE54993 | LPCAT1 | 79888 | 5 | 5 | 60 | |
| GSE54994 | LPCAT1 | 79888 | 29 | 1 | 23 | |
| GSE60625 | LPCAT1 | 79888 | 0 | 0 | 11 | |
| GSE74703 | LPCAT1 | 79888 | 14 | 0 | 22 | |
| GSE74704 | LPCAT1 | 79888 | 11 | 0 | 9 | |
| TCGA | LPCAT1 | 79888 | 59 | 3 | 34 |
Total number of gains: 174; Total number of losses: 45; Total Number of normals: 269.
Somatic mutations of LPCAT1:
Generating mutation plots.
Highly correlated genes for LPCAT1:
Showing top 20/2336 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LPCAT1 | FMNL2 | 0.867226 | 7 | 0 | 7 |
| LPCAT1 | FADS1 | 0.843774 | 3 | 0 | 3 |
| LPCAT1 | SERPINH1 | 0.838799 | 10 | 0 | 10 |
| LPCAT1 | FNDC3B | 0.838559 | 11 | 0 | 11 |
| LPCAT1 | PLOD1 | 0.822481 | 12 | 0 | 12 |
| LPCAT1 | MSN | 0.819777 | 11 | 0 | 11 |
| LPCAT1 | WDR54 | 0.817129 | 7 | 0 | 7 |
| LPCAT1 | XPR1 | 0.814452 | 7 | 0 | 7 |
| LPCAT1 | GNG10 | 0.811749 | 3 | 0 | 3 |
| LPCAT1 | CTHRC1 | 0.808197 | 7 | 0 | 6 |
| LPCAT1 | LRRC8C | 0.802025 | 3 | 0 | 3 |
| LPCAT1 | COLGALT1 | 0.799878 | 10 | 0 | 10 |
| LPCAT1 | LAPTM4B | 0.799768 | 11 | 0 | 11 |
| LPCAT1 | LRP12 | 0.798764 | 11 | 0 | 11 |
| LPCAT1 | TRAM2 | 0.798677 | 11 | 0 | 11 |
| LPCAT1 | CCDC127 | 0.7978 | 3 | 0 | 3 |
| LPCAT1 | TMEM184B | 0.793441 | 11 | 0 | 11 |
| LPCAT1 | SLC39A14 | 0.787468 | 11 | 0 | 11 |
| LPCAT1 | RAB34 | 0.786185 | 3 | 0 | 3 |
| LPCAT1 | SDHAF2 | 0.78594 | 3 | 0 | 3 |
For details and further investigation, click here