| Full name: tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein eta | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 22q12.3 | ||
| Entrez ID: 7533 | HGNC ID: HGNC:12853 | Ensembl Gene: ENSG00000128245 | OMIM ID: 113508 |
| Drug and gene relationship at DGIdb | |||
YWHAH involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04110 | Cell cycle | |
| hsa04114 | Oocyte meiosis | |
| hsa04151 | PI3K-Akt signaling pathway | |
| hsa04390 | Hippo signaling pathway |
Expression of YWHAH:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | YWHAH | 7533 | 201020_at | 0.5445 | 0.3194 | |
| GSE20347 | YWHAH | 7533 | 201020_at | 0.6508 | 0.0030 | |
| GSE23400 | YWHAH | 7533 | 201020_at | 0.8049 | 0.0000 | |
| GSE26886 | YWHAH | 7533 | 201020_at | -0.8887 | 0.0004 | |
| GSE29001 | YWHAH | 7533 | 201020_at | 0.6416 | 0.0048 | |
| GSE38129 | YWHAH | 7533 | 201020_at | 0.6926 | 0.0173 | |
| GSE45670 | YWHAH | 7533 | 201020_at | 0.4660 | 0.0186 | |
| GSE53622 | YWHAH | 7533 | 97714 | 0.7271 | 0.0000 | |
| GSE53624 | YWHAH | 7533 | 97714 | 0.8356 | 0.0000 | |
| GSE63941 | YWHAH | 7533 | 201020_at | -0.3129 | 0.5274 | |
| GSE77861 | YWHAH | 7533 | 201020_at | 1.0972 | 0.0057 | |
| GSE97050 | YWHAH | 7533 | A_23_P103070 | 0.5097 | 0.2056 | |
| SRP007169 | YWHAH | 7533 | RNAseq | 1.1768 | 0.0001 | |
| SRP008496 | YWHAH | 7533 | RNAseq | 1.3244 | 0.0000 | |
| SRP064894 | YWHAH | 7533 | RNAseq | 1.0099 | 0.0000 | |
| SRP133303 | YWHAH | 7533 | RNAseq | 0.9524 | 0.0000 | |
| SRP159526 | YWHAH | 7533 | RNAseq | 0.3533 | 0.2246 | |
| SRP193095 | YWHAH | 7533 | RNAseq | 0.1689 | 0.3920 | |
| SRP219564 | YWHAH | 7533 | RNAseq | 0.2944 | 0.5770 | |
| TCGA | YWHAH | 7533 | RNAseq | 0.0301 | 0.4854 |
Upregulated datasets: 4; Downregulated datasets: 0.
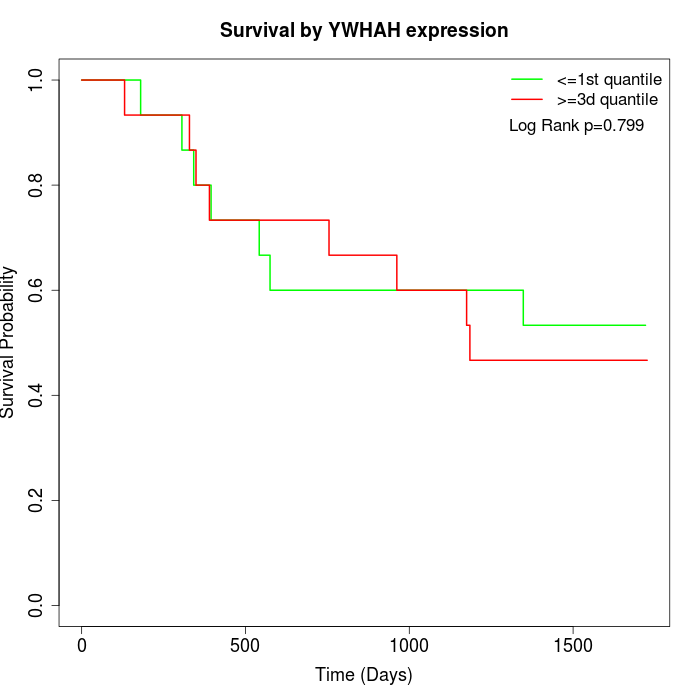
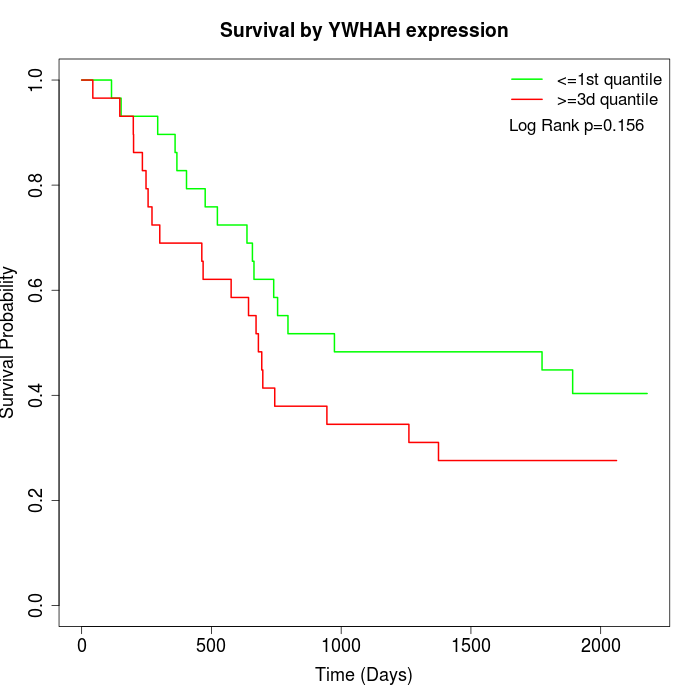
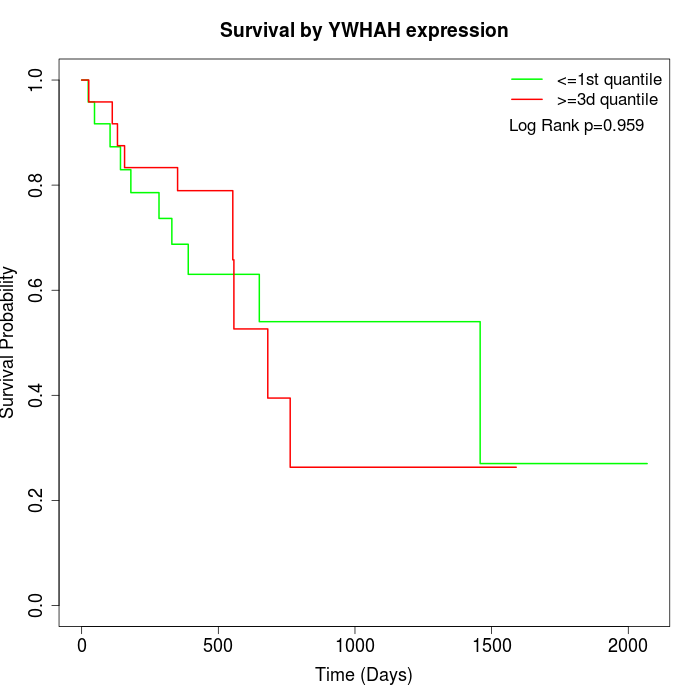
Survival by YWHAH expression:
Note: Click image to view full size file.
Copy number change of YWHAH:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | YWHAH | 7533 | 4 | 4 | 22 | |
| GSE20123 | YWHAH | 7533 | 4 | 3 | 23 | |
| GSE43470 | YWHAH | 7533 | 4 | 5 | 34 | |
| GSE46452 | YWHAH | 7533 | 31 | 1 | 27 | |
| GSE47630 | YWHAH | 7533 | 8 | 5 | 27 | |
| GSE54993 | YWHAH | 7533 | 3 | 6 | 61 | |
| GSE54994 | YWHAH | 7533 | 9 | 9 | 35 | |
| GSE60625 | YWHAH | 7533 | 5 | 0 | 6 | |
| GSE74703 | YWHAH | 7533 | 4 | 5 | 27 | |
| GSE74704 | YWHAH | 7533 | 1 | 1 | 18 | |
| TCGA | YWHAH | 7533 | 28 | 16 | 52 |
Total number of gains: 101; Total number of losses: 55; Total Number of normals: 332.
Somatic mutations of YWHAH:
Generating mutation plots.
Highly correlated genes for YWHAH:
Showing top 20/1005 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| YWHAH | LRRC8C | 0.802397 | 3 | 0 | 3 |
| YWHAH | FADS1 | 0.7934 | 3 | 0 | 3 |
| YWHAH | RAB34 | 0.79259 | 3 | 0 | 3 |
| YWHAH | PYCR2 | 0.752111 | 3 | 0 | 3 |
| YWHAH | CERS2 | 0.74654 | 8 | 0 | 8 |
| YWHAH | TMTC1 | 0.739417 | 3 | 0 | 3 |
| YWHAH | GNG10 | 0.736044 | 3 | 0 | 3 |
| YWHAH | KLHL5 | 0.734708 | 6 | 0 | 6 |
| YWHAH | PDIA5 | 0.729649 | 3 | 0 | 3 |
| YWHAH | IFITM3 | 0.72311 | 10 | 0 | 10 |
| YWHAH | NF1 | 0.722766 | 3 | 0 | 3 |
| YWHAH | FUCA2 | 0.720311 | 5 | 0 | 5 |
| YWHAH | CALU | 0.717638 | 13 | 0 | 10 |
| YWHAH | FCGR3A | 0.71065 | 3 | 0 | 3 |
| YWHAH | TXNDC12 | 0.708384 | 4 | 0 | 4 |
| YWHAH | HNRNPU | 0.702603 | 9 | 0 | 9 |
| YWHAH | MSN | 0.701703 | 10 | 0 | 9 |
| YWHAH | SLC35B2 | 0.696234 | 3 | 0 | 3 |
| YWHAH | MB21D2 | 0.693029 | 5 | 0 | 5 |
| YWHAH | PARVB | 0.692438 | 9 | 0 | 8 |
For details and further investigation, click here