| Full name: Fc receptor like A | Alias Symbol: MGC4595|FCRLc2|FCRLb|FCRLc1|FCRLd|FCRLe|FCRL|FCRLa|FREB|FCRLX | ||
| Type: protein-coding gene | Cytoband: 1q23.3 | ||
| Entrez ID: 84824 | HGNC ID: HGNC:18504 | Ensembl Gene: ENSG00000132185 | OMIM ID: 606891 |
| Drug and gene relationship at DGIdb | |||
Expression of FCRLA:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | FCRLA | 84824 | 235372_at | -0.1911 | 0.8722 | |
| GSE26886 | FCRLA | 84824 | 235372_at | -0.2539 | 0.1856 | |
| GSE45670 | FCRLA | 84824 | 235372_at | -0.1376 | 0.5238 | |
| GSE53622 | FCRLA | 84824 | 48518 | -0.7847 | 0.0058 | |
| GSE53624 | FCRLA | 84824 | 48518 | -1.2954 | 0.0000 | |
| GSE63941 | FCRLA | 84824 | 235372_at | -0.1879 | 0.5380 | |
| GSE77861 | FCRLA | 84824 | 235372_at | -0.4211 | 0.0057 | |
| GSE97050 | FCRLA | 84824 | A_23_P46039 | 0.4936 | 0.1992 | |
| SRP133303 | FCRLA | 84824 | RNAseq | -0.2638 | 0.1824 | |
| SRP159526 | FCRLA | 84824 | RNAseq | -2.1821 | 0.0316 | |
| TCGA | FCRLA | 84824 | RNAseq | -0.6282 | 0.2227 |
Upregulated datasets: 0; Downregulated datasets: 2.
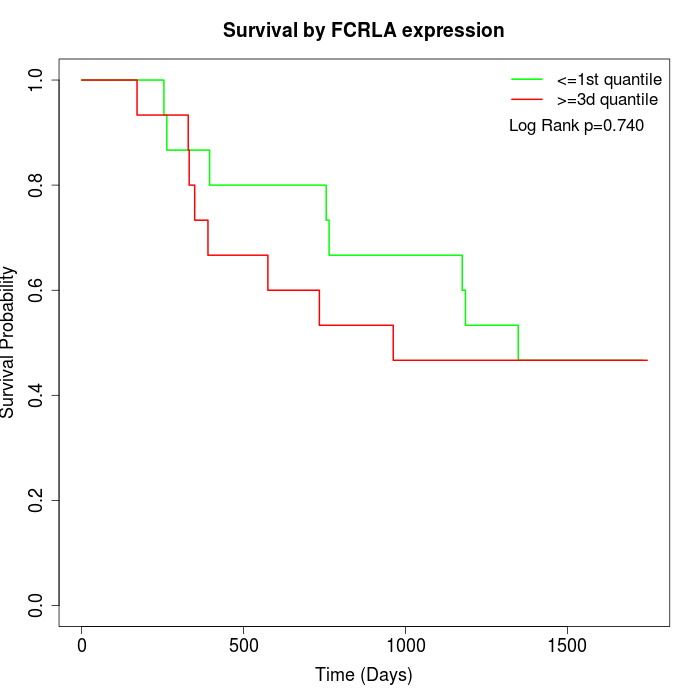
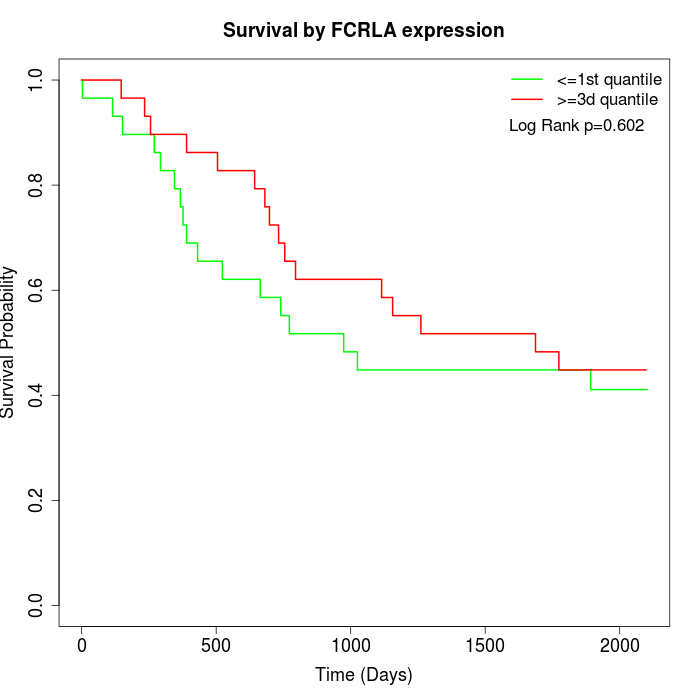
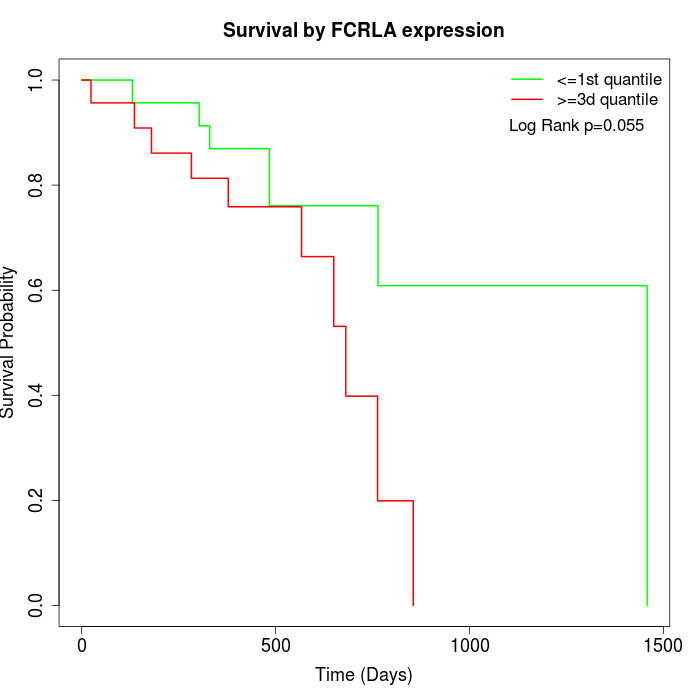
Survival by FCRLA expression:
Note: Click image to view full size file.
Copy number change of FCRLA:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | FCRLA | 84824 | 10 | 0 | 20 | |
| GSE20123 | FCRLA | 84824 | 10 | 0 | 20 | |
| GSE43470 | FCRLA | 84824 | 7 | 2 | 34 | |
| GSE46452 | FCRLA | 84824 | 2 | 1 | 56 | |
| GSE47630 | FCRLA | 84824 | 14 | 0 | 26 | |
| GSE54993 | FCRLA | 84824 | 0 | 5 | 65 | |
| GSE54994 | FCRLA | 84824 | 16 | 0 | 37 | |
| GSE60625 | FCRLA | 84824 | 0 | 0 | 11 | |
| GSE74703 | FCRLA | 84824 | 7 | 2 | 27 | |
| GSE74704 | FCRLA | 84824 | 4 | 0 | 16 | |
| TCGA | FCRLA | 84824 | 43 | 4 | 49 |
Total number of gains: 113; Total number of losses: 14; Total Number of normals: 361.
Somatic mutations of FCRLA:
Generating mutation plots.
Highly correlated genes for FCRLA:
Showing top 20/281 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| FCRLA | CD79B | 0.841271 | 5 | 0 | 5 |
| FCRLA | CD37 | 0.825761 | 6 | 0 | 6 |
| FCRLA | MS4A1 | 0.809244 | 6 | 0 | 6 |
| FCRLA | FCRL3 | 0.808499 | 7 | 0 | 7 |
| FCRLA | CLEC17A | 0.788568 | 3 | 0 | 3 |
| FCRLA | STAP1 | 0.757217 | 6 | 0 | 5 |
| FCRLA | TAGAP | 0.75135 | 4 | 0 | 4 |
| FCRLA | BTLA | 0.724028 | 6 | 0 | 5 |
| FCRLA | PNOC | 0.722031 | 4 | 0 | 4 |
| FCRLA | RNF208 | 0.718447 | 3 | 0 | 3 |
| FCRLA | KCNA3 | 0.714833 | 5 | 0 | 4 |
| FCRLA | CD48 | 0.714512 | 5 | 0 | 4 |
| FCRLA | POU2AF1 | 0.714367 | 4 | 0 | 4 |
| FCRLA | ETS1 | 0.714219 | 3 | 0 | 3 |
| FCRLA | IRF4 | 0.709032 | 6 | 0 | 5 |
| FCRLA | LINC01215 | 0.70458 | 5 | 0 | 4 |
| FCRLA | IKZF3 | 0.701891 | 5 | 0 | 5 |
| FCRLA | MUL1 | 0.696819 | 3 | 0 | 3 |
| FCRLA | FMN1 | 0.695866 | 4 | 0 | 4 |
| FCRLA | DPEP2 | 0.693081 | 5 | 0 | 5 |
For details and further investigation, click here