| Full name: potassium voltage-gated channel subfamily A member 3 | Alias Symbol: Kv1.3|MK3|HLK3|HPCN3 | ||
| Type: protein-coding gene | Cytoband: 1p13.3 | ||
| Entrez ID: 3738 | HGNC ID: HGNC:6221 | Ensembl Gene: ENSG00000177272 | OMIM ID: 176263 |
| Drug and gene relationship at DGIdb | |||
Expression of KCNA3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCNA3 | 3738 | 207237_at | 0.0425 | 0.9359 | |
| GSE20347 | KCNA3 | 3738 | 207237_at | 0.0473 | 0.4829 | |
| GSE23400 | KCNA3 | 3738 | 207237_at | -0.1042 | 0.0014 | |
| GSE26886 | KCNA3 | 3738 | 207237_at | -0.2064 | 0.0756 | |
| GSE29001 | KCNA3 | 3738 | 207237_at | -0.0568 | 0.6610 | |
| GSE38129 | KCNA3 | 3738 | 207237_at | -0.1209 | 0.1299 | |
| GSE45670 | KCNA3 | 3738 | 207237_at | -0.1424 | 0.0413 | |
| GSE53622 | KCNA3 | 3738 | 47011 | 0.0191 | 0.9355 | |
| GSE53624 | KCNA3 | 3738 | 47011 | -0.4096 | 0.0411 | |
| GSE63941 | KCNA3 | 3738 | 207237_at | 0.1075 | 0.3416 | |
| GSE77861 | KCNA3 | 3738 | 207237_at | -0.0130 | 0.9264 | |
| GSE97050 | KCNA3 | 3738 | A_33_P3415032 | 1.5455 | 0.1239 | |
| SRP007169 | KCNA3 | 3738 | RNAseq | 0.5349 | 0.4468 | |
| SRP064894 | KCNA3 | 3738 | RNAseq | 0.0352 | 0.9237 | |
| SRP133303 | KCNA3 | 3738 | RNAseq | -0.3819 | 0.2875 | |
| SRP159526 | KCNA3 | 3738 | RNAseq | -0.2032 | 0.8285 | |
| SRP193095 | KCNA3 | 3738 | RNAseq | -0.1608 | 0.6705 | |
| SRP219564 | KCNA3 | 3738 | RNAseq | 0.1655 | 0.7978 | |
| TCGA | KCNA3 | 3738 | RNAseq | -0.8019 | 0.0133 |
Upregulated datasets: 0; Downregulated datasets: 0.
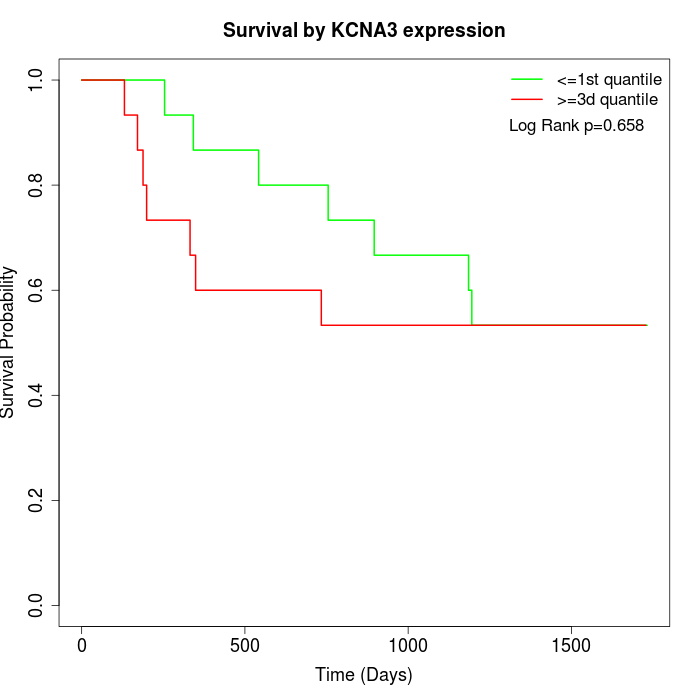
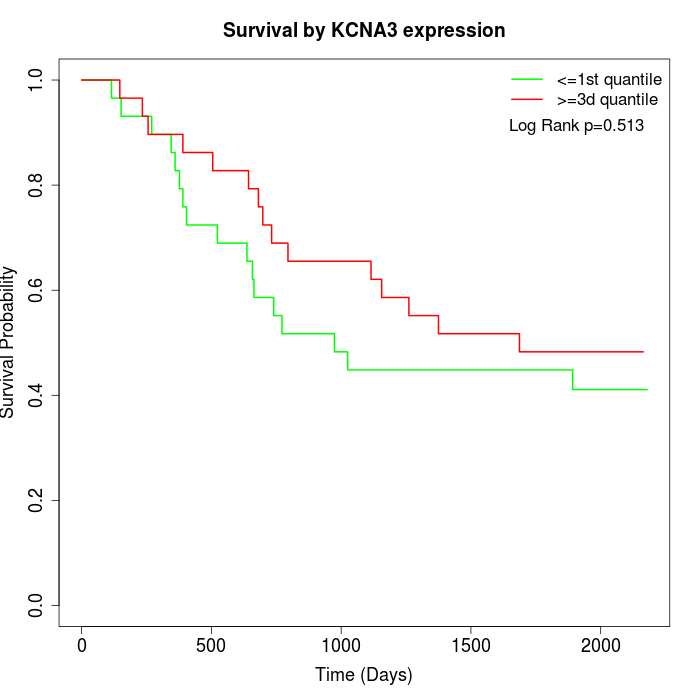
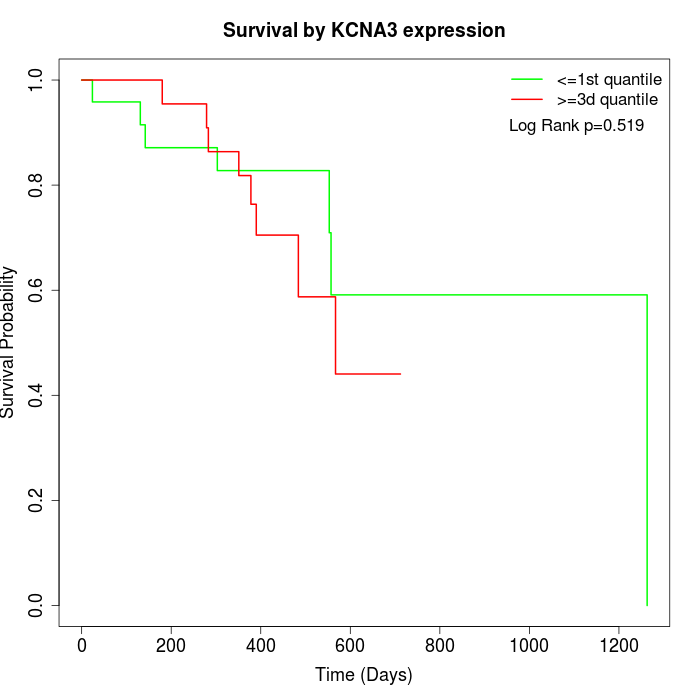
Survival by KCNA3 expression:
Note: Click image to view full size file.
Copy number change of KCNA3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCNA3 | 3738 | 0 | 11 | 19 | |
| GSE20123 | KCNA3 | 3738 | 0 | 11 | 19 | |
| GSE43470 | KCNA3 | 3738 | 0 | 8 | 35 | |
| GSE46452 | KCNA3 | 3738 | 2 | 1 | 56 | |
| GSE47630 | KCNA3 | 3738 | 9 | 5 | 26 | |
| GSE54993 | KCNA3 | 3738 | 0 | 1 | 69 | |
| GSE54994 | KCNA3 | 3738 | 7 | 4 | 42 | |
| GSE60625 | KCNA3 | 3738 | 0 | 0 | 11 | |
| GSE74703 | KCNA3 | 3738 | 0 | 7 | 29 | |
| GSE74704 | KCNA3 | 3738 | 0 | 7 | 13 | |
| TCGA | KCNA3 | 3738 | 8 | 27 | 61 |
Total number of gains: 26; Total number of losses: 82; Total Number of normals: 380.
Somatic mutations of KCNA3:
Generating mutation plots.
Highly correlated genes for KCNA3:
Showing top 20/491 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCNA3 | POU2AF1 | 0.84627 | 3 | 0 | 3 |
| KCNA3 | HVCN1 | 0.792007 | 5 | 0 | 5 |
| KCNA3 | PARVG | 0.764185 | 4 | 0 | 4 |
| KCNA3 | SUSD3 | 0.74763 | 4 | 0 | 4 |
| KCNA3 | TNFSF8 | 0.740038 | 7 | 0 | 7 |
| KCNA3 | FCRL5 | 0.735347 | 4 | 0 | 4 |
| KCNA3 | EVI2B | 0.734521 | 6 | 0 | 6 |
| KCNA3 | PIP4K2A | 0.729104 | 3 | 0 | 3 |
| KCNA3 | TIFAB | 0.725703 | 3 | 0 | 3 |
| KCNA3 | TAGAP | 0.72503 | 5 | 0 | 4 |
| KCNA3 | FCRL3 | 0.724354 | 5 | 0 | 4 |
| KCNA3 | FCRLA | 0.714833 | 5 | 0 | 4 |
| KCNA3 | CLEC17A | 0.709318 | 3 | 0 | 3 |
| KCNA3 | GIMAP5 | 0.707192 | 3 | 0 | 3 |
| KCNA3 | TIGIT | 0.70692 | 4 | 0 | 4 |
| KCNA3 | HLA-DMB | 0.70674 | 5 | 0 | 5 |
| KCNA3 | IL10RA | 0.702985 | 4 | 0 | 4 |
| KCNA3 | PSMB8-AS1 | 0.701303 | 3 | 0 | 3 |
| KCNA3 | EAF2 | 0.700435 | 4 | 0 | 4 |
| KCNA3 | PIK3R6 | 0.693728 | 4 | 0 | 3 |
For details and further investigation, click here