| Full name: C-C motif chemokine ligand 28 | Alias Symbol: SCYA28|MEC|CCK1 | ||
| Type: protein-coding gene | Cytoband: 5p12 | ||
| Entrez ID: 56477 | HGNC ID: HGNC:17700 | Ensembl Gene: ENSG00000151882 | OMIM ID: 605240 |
| Drug and gene relationship at DGIdb | |||
CCL28 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04062 | Chemokine signaling pathway |
Expression of CCL28:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CCL28 | 56477 | 238750_at | -0.3758 | 0.7131 | |
| GSE26886 | CCL28 | 56477 | 224240_s_at | 0.3260 | 0.1966 | |
| GSE45670 | CCL28 | 56477 | 238750_at | -0.5245 | 0.0518 | |
| GSE53622 | CCL28 | 56477 | 74561 | -0.9372 | 0.0000 | |
| GSE53624 | CCL28 | 56477 | 74561 | -0.9362 | 0.0000 | |
| GSE63941 | CCL28 | 56477 | 238750_at | 0.9774 | 0.4684 | |
| GSE77861 | CCL28 | 56477 | 238750_at | -0.2998 | 0.0447 | |
| GSE97050 | CCL28 | 56477 | A_23_P503072 | -0.1473 | 0.8156 | |
| SRP007169 | CCL28 | 56477 | RNAseq | 1.4137 | 0.1142 | |
| SRP064894 | CCL28 | 56477 | RNAseq | -1.0471 | 0.0040 | |
| SRP133303 | CCL28 | 56477 | RNAseq | -0.7003 | 0.0049 | |
| SRP159526 | CCL28 | 56477 | RNAseq | -0.2339 | 0.7569 | |
| SRP193095 | CCL28 | 56477 | RNAseq | -0.6581 | 0.0132 | |
| SRP219564 | CCL28 | 56477 | RNAseq | -0.7980 | 0.3083 | |
| TCGA | CCL28 | 56477 | RNAseq | -0.4965 | 0.0553 |
Upregulated datasets: 0; Downregulated datasets: 1.
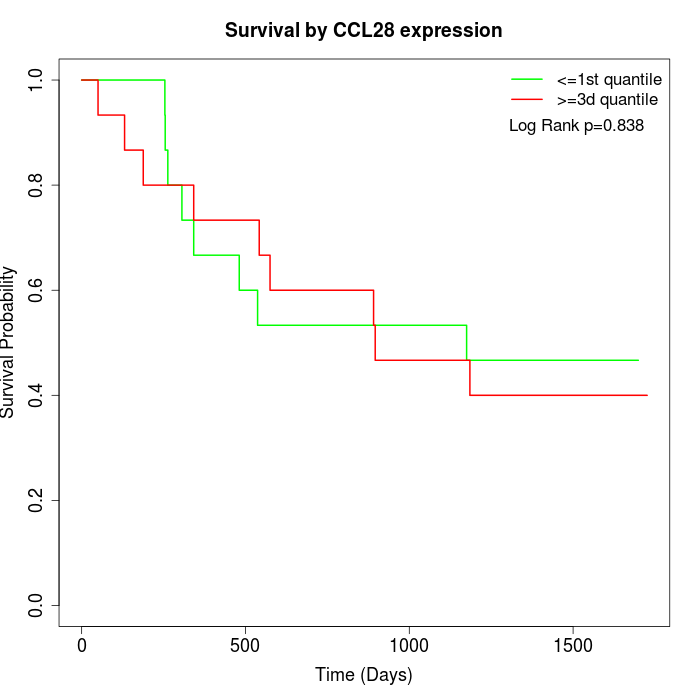
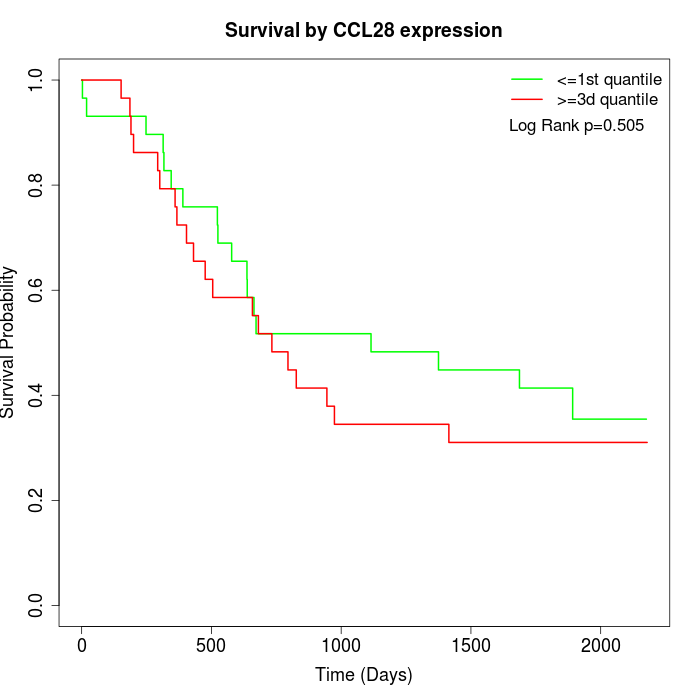
Survival by CCL28 expression:
Note: Click image to view full size file.
Copy number change of CCL28:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CCL28 | 56477 | 10 | 0 | 20 | |
| GSE20123 | CCL28 | 56477 | 10 | 0 | 20 | |
| GSE43470 | CCL28 | 56477 | 17 | 0 | 26 | |
| GSE46452 | CCL28 | 56477 | 5 | 22 | 32 | |
| GSE47630 | CCL28 | 56477 | 6 | 12 | 22 | |
| GSE54993 | CCL28 | 56477 | 6 | 3 | 61 | |
| GSE54994 | CCL28 | 56477 | 24 | 2 | 27 | |
| GSE60625 | CCL28 | 56477 | 0 | 0 | 11 | |
| GSE74703 | CCL28 | 56477 | 13 | 0 | 23 | |
| GSE74704 | CCL28 | 56477 | 9 | 0 | 11 | |
| TCGA | CCL28 | 56477 | 53 | 6 | 37 |
Total number of gains: 153; Total number of losses: 45; Total Number of normals: 290.
Somatic mutations of CCL28:
Generating mutation plots.
Highly correlated genes for CCL28:
Showing top 20/115 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CCL28 | FCGBP | 0.797547 | 3 | 0 | 3 |
| CCL28 | ZNF175 | 0.754542 | 3 | 0 | 3 |
| CCL28 | MAT2B | 0.716472 | 3 | 0 | 3 |
| CCL28 | NCOA7 | 0.714952 | 3 | 0 | 3 |
| CCL28 | B4GALT5 | 0.705749 | 3 | 0 | 3 |
| CCL28 | SURF1 | 0.694612 | 3 | 0 | 3 |
| CCL28 | LRG1 | 0.690999 | 3 | 0 | 3 |
| CCL28 | APIP | 0.685813 | 3 | 0 | 3 |
| CCL28 | AKAP7 | 0.67954 | 3 | 0 | 3 |
| CCL28 | TRERF1 | 0.67909 | 3 | 0 | 3 |
| CCL28 | PKIA | 0.677239 | 3 | 0 | 3 |
| CCL28 | TAF12 | 0.675275 | 3 | 0 | 3 |
| CCL28 | LMO4 | 0.668626 | 3 | 0 | 3 |
| CCL28 | PPM1K | 0.667782 | 3 | 0 | 3 |
| CCL28 | BIRC3 | 0.667623 | 4 | 0 | 3 |
| CCL28 | HLA-DQB1 | 0.666088 | 3 | 0 | 3 |
| CCL28 | EPS15 | 0.663536 | 3 | 0 | 3 |
| CCL28 | PVRIG | 0.66215 | 3 | 0 | 3 |
| CCL28 | DRAM2 | 0.662102 | 3 | 0 | 3 |
| CCL28 | HLA-DRA | 0.661607 | 3 | 0 | 3 |
For details and further investigation, click here