| Full name: FRAT regulator of WNT signaling pathway 2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 10q24.1 | ||
| Entrez ID: 23401 | HGNC ID: HGNC:16048 | Ensembl Gene: ENSG00000181274 | OMIM ID: 605006 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
FRAT2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04310 | Wnt signaling pathway |
Expression of FRAT2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | FRAT2 | 23401 | 209864_at | 0.4180 | 0.5568 | |
| GSE20347 | FRAT2 | 23401 | 209864_at | -0.0737 | 0.7010 | |
| GSE23400 | FRAT2 | 23401 | 209864_at | 0.1873 | 0.0288 | |
| GSE26886 | FRAT2 | 23401 | 209864_at | -0.9831 | 0.0084 | |
| GSE29001 | FRAT2 | 23401 | 209864_at | 0.0220 | 0.9630 | |
| GSE38129 | FRAT2 | 23401 | 209864_at | 0.0988 | 0.5553 | |
| GSE45670 | FRAT2 | 23401 | 209864_at | -0.0713 | 0.7827 | |
| GSE53622 | FRAT2 | 23401 | 9171 | -0.3019 | 0.0072 | |
| GSE53624 | FRAT2 | 23401 | 9171 | -0.1247 | 0.1513 | |
| GSE63941 | FRAT2 | 23401 | 209864_at | 1.1609 | 0.0636 | |
| GSE77861 | FRAT2 | 23401 | 209864_at | -0.1412 | 0.7417 | |
| GSE97050 | FRAT2 | 23401 | A_23_P12784 | -0.1026 | 0.5752 | |
| SRP007169 | FRAT2 | 23401 | RNAseq | -1.0939 | 0.0738 | |
| SRP008496 | FRAT2 | 23401 | RNAseq | -0.9264 | 0.0095 | |
| SRP064894 | FRAT2 | 23401 | RNAseq | -0.2520 | 0.2626 | |
| SRP133303 | FRAT2 | 23401 | RNAseq | -0.3206 | 0.2384 | |
| SRP159526 | FRAT2 | 23401 | RNAseq | 0.2147 | 0.6630 | |
| SRP193095 | FRAT2 | 23401 | RNAseq | -0.5365 | 0.0036 | |
| SRP219564 | FRAT2 | 23401 | RNAseq | 0.9537 | 0.1957 | |
| TCGA | FRAT2 | 23401 | RNAseq | 0.1178 | 0.1976 |
Upregulated datasets: 0; Downregulated datasets: 0.
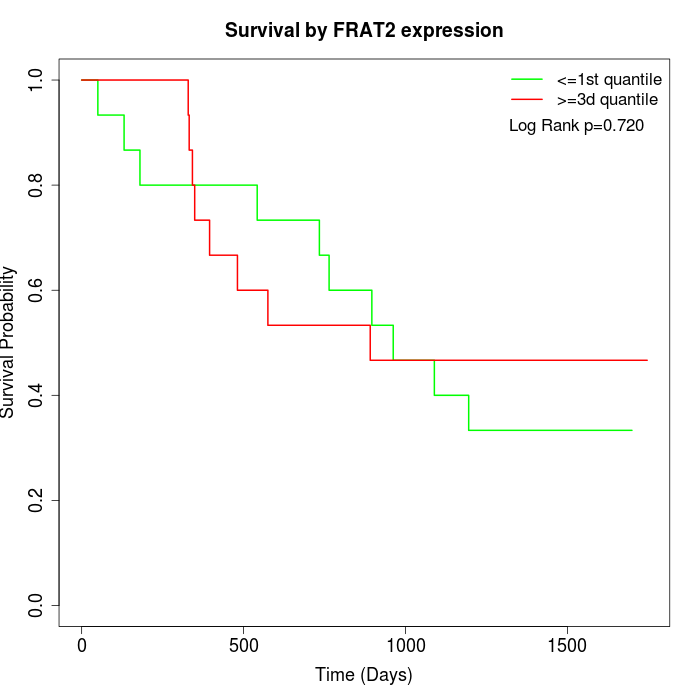
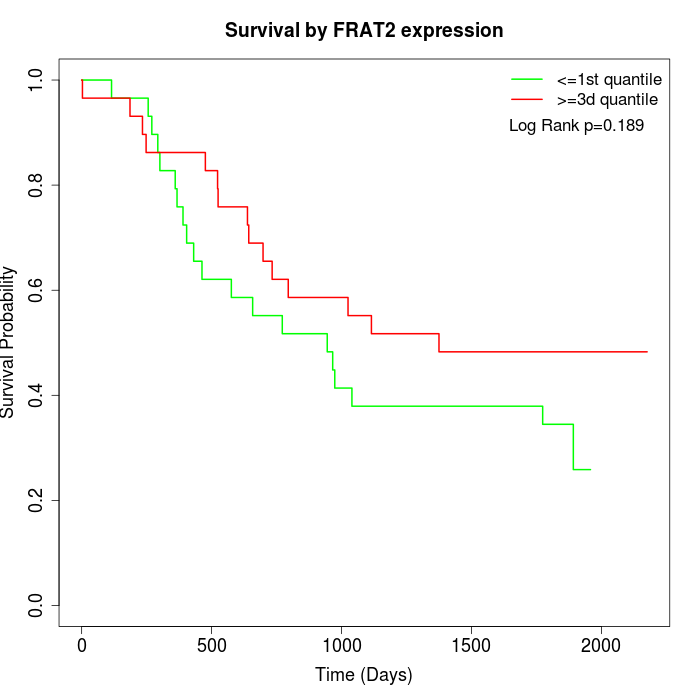
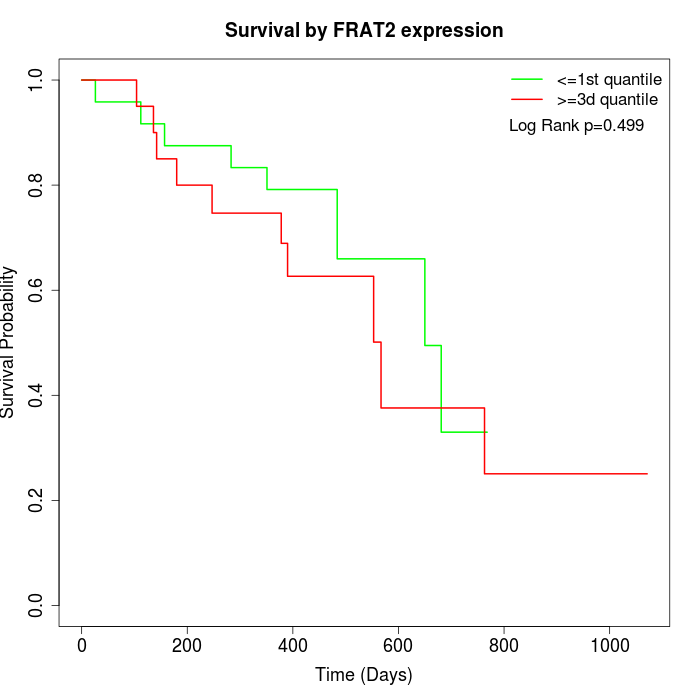
Survival by FRAT2 expression:
Note: Click image to view full size file.
Copy number change of FRAT2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | FRAT2 | 23401 | 0 | 8 | 22 | |
| GSE20123 | FRAT2 | 23401 | 0 | 7 | 23 | |
| GSE43470 | FRAT2 | 23401 | 0 | 7 | 36 | |
| GSE46452 | FRAT2 | 23401 | 0 | 11 | 48 | |
| GSE47630 | FRAT2 | 23401 | 2 | 14 | 24 | |
| GSE54993 | FRAT2 | 23401 | 8 | 0 | 62 | |
| GSE54994 | FRAT2 | 23401 | 2 | 10 | 41 | |
| GSE60625 | FRAT2 | 23401 | 0 | 0 | 11 | |
| GSE74703 | FRAT2 | 23401 | 0 | 5 | 31 | |
| GSE74704 | FRAT2 | 23401 | 0 | 4 | 16 | |
| TCGA | FRAT2 | 23401 | 5 | 28 | 63 |
Total number of gains: 17; Total number of losses: 94; Total Number of normals: 377.
Somatic mutations of FRAT2:
Generating mutation plots.
Highly correlated genes for FRAT2:
Showing top 20/393 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| FRAT2 | COMMD2 | 0.754587 | 3 | 0 | 3 |
| FRAT2 | ZNF606 | 0.727379 | 3 | 0 | 3 |
| FRAT2 | ELOF1 | 0.72187 | 3 | 0 | 3 |
| FRAT2 | UBE2N | 0.717081 | 3 | 0 | 3 |
| FRAT2 | HIC2 | 0.693679 | 3 | 0 | 3 |
| FRAT2 | GABPA | 0.691123 | 3 | 0 | 3 |
| FRAT2 | DCTD | 0.690104 | 4 | 0 | 3 |
| FRAT2 | ZFPL1 | 0.683117 | 3 | 0 | 3 |
| FRAT2 | MDN1 | 0.677366 | 3 | 0 | 3 |
| FRAT2 | RCAN3 | 0.67209 | 5 | 0 | 3 |
| FRAT2 | FGGY | 0.671209 | 5 | 0 | 4 |
| FRAT2 | CWF19L1 | 0.670977 | 4 | 0 | 3 |
| FRAT2 | USP1 | 0.661338 | 4 | 0 | 3 |
| FRAT2 | TXNDC16 | 0.657854 | 4 | 0 | 3 |
| FRAT2 | PPP1R8 | 0.656857 | 4 | 0 | 3 |
| FRAT2 | ARFGAP2 | 0.656209 | 4 | 0 | 4 |
| FRAT2 | EXD2 | 0.652515 | 3 | 0 | 3 |
| FRAT2 | ARAF | 0.652511 | 3 | 0 | 3 |
| FRAT2 | SLC44A3 | 0.649859 | 3 | 0 | 3 |
| FRAT2 | CRNKL1 | 0.646729 | 4 | 0 | 3 |
For details and further investigation, click here