| Full name: fibroblast growth factor receptor substrate 2 | Alias Symbol: SNT-1|FRS2alpha|SNT1|FRS2A | ||
| Type: protein-coding gene | Cytoband: 12q15 | ||
| Entrez ID: 10818 | HGNC ID: HGNC:16971 | Ensembl Gene: ENSG00000166225 | OMIM ID: 607743 |
| Drug and gene relationship at DGIdb | |||
FRS2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04722 | Neurotrophin signaling pathway | |
| hsa05205 | Proteoglycans in cancer |
Expression of FRS2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | FRS2 | 10818 | 226045_at | 0.1210 | 0.8587 | |
| GSE20347 | FRS2 | 10818 | 221308_at | -0.0839 | 0.2787 | |
| GSE23400 | FRS2 | 10818 | 221308_at | -0.1879 | 0.0000 | |
| GSE26886 | FRS2 | 10818 | 226045_at | 0.1851 | 0.6430 | |
| GSE29001 | FRS2 | 10818 | 221308_at | -0.1255 | 0.1751 | |
| GSE38129 | FRS2 | 10818 | 221308_at | -0.1332 | 0.1170 | |
| GSE45670 | FRS2 | 10818 | 226045_at | -0.1522 | 0.4035 | |
| GSE53622 | FRS2 | 10818 | 149862 | -0.4674 | 0.0000 | |
| GSE53624 | FRS2 | 10818 | 149862 | -0.1005 | 0.2719 | |
| GSE63941 | FRS2 | 10818 | 226045_at | -0.8073 | 0.3802 | |
| GSE77861 | FRS2 | 10818 | 226045_at | 0.1146 | 0.7185 | |
| GSE97050 | FRS2 | 10818 | A_33_P3364112 | -0.2827 | 0.2850 | |
| SRP007169 | FRS2 | 10818 | RNAseq | 0.1161 | 0.7949 | |
| SRP008496 | FRS2 | 10818 | RNAseq | 0.2683 | 0.2503 | |
| SRP064894 | FRS2 | 10818 | RNAseq | 0.0553 | 0.8194 | |
| SRP133303 | FRS2 | 10818 | RNAseq | 0.3124 | 0.0048 | |
| SRP159526 | FRS2 | 10818 | RNAseq | 0.2792 | 0.3445 | |
| SRP193095 | FRS2 | 10818 | RNAseq | 0.2056 | 0.0849 | |
| TCGA | FRS2 | 10818 | RNAseq | 0.0093 | 0.8912 |
Upregulated datasets: 0; Downregulated datasets: 0.
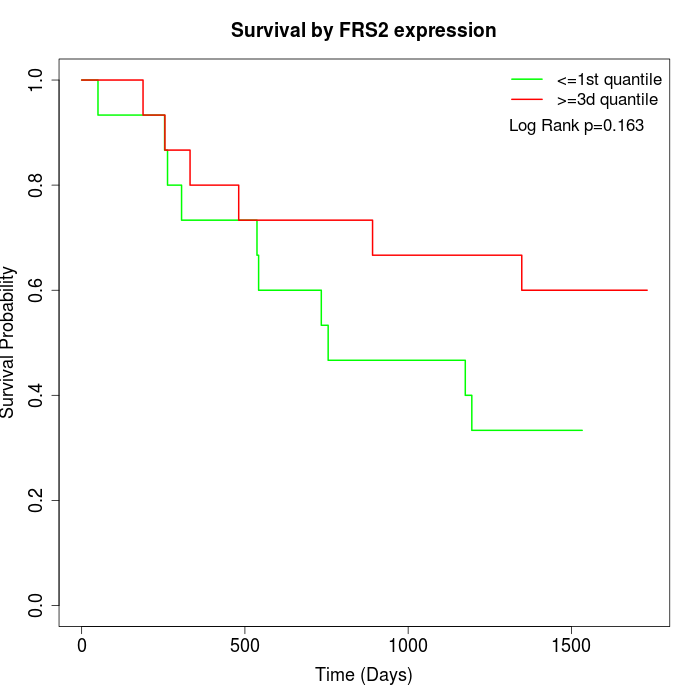
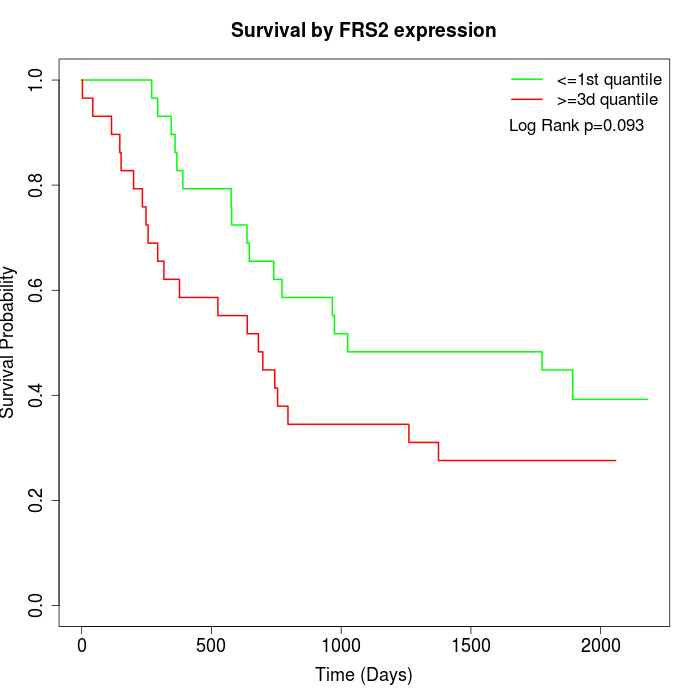
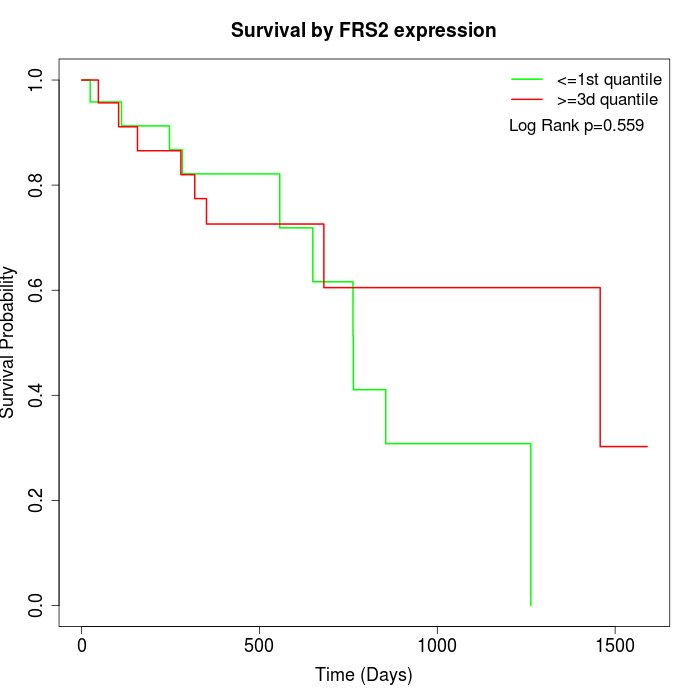
Survival by FRS2 expression:
Note: Click image to view full size file.
Copy number change of FRS2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | FRS2 | 10818 | 6 | 1 | 23 | |
| GSE20123 | FRS2 | 10818 | 6 | 0 | 24 | |
| GSE43470 | FRS2 | 10818 | 4 | 0 | 39 | |
| GSE46452 | FRS2 | 10818 | 11 | 1 | 47 | |
| GSE47630 | FRS2 | 10818 | 10 | 1 | 29 | |
| GSE54993 | FRS2 | 10818 | 0 | 6 | 64 | |
| GSE54994 | FRS2 | 10818 | 8 | 1 | 44 | |
| GSE60625 | FRS2 | 10818 | 0 | 0 | 11 | |
| GSE74703 | FRS2 | 10818 | 4 | 0 | 32 | |
| GSE74704 | FRS2 | 10818 | 5 | 0 | 15 | |
| TCGA | FRS2 | 10818 | 23 | 8 | 65 |
Total number of gains: 77; Total number of losses: 18; Total Number of normals: 393.
Somatic mutations of FRS2:
Generating mutation plots.
Highly correlated genes for FRS2:
Showing top 20/286 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| FRS2 | ABCB10 | 0.808263 | 3 | 0 | 3 |
| FRS2 | INTS10 | 0.787634 | 3 | 0 | 3 |
| FRS2 | RTN4IP1 | 0.767682 | 3 | 0 | 3 |
| FRS2 | EIF2AK4 | 0.76114 | 3 | 0 | 3 |
| FRS2 | TET2 | 0.758603 | 3 | 0 | 3 |
| FRS2 | PRMT6 | 0.754405 | 3 | 0 | 3 |
| FRS2 | TRIAP1 | 0.740075 | 3 | 0 | 3 |
| FRS2 | COX5A | 0.731794 | 3 | 0 | 3 |
| FRS2 | PTPN13 | 0.72894 | 3 | 0 | 3 |
| FRS2 | KANSL2 | 0.719988 | 3 | 0 | 3 |
| FRS2 | TXNDC16 | 0.716895 | 3 | 0 | 3 |
| FRS2 | C6orf120 | 0.709065 | 5 | 0 | 4 |
| FRS2 | ZNF260 | 0.700891 | 3 | 0 | 3 |
| FRS2 | OCRL | 0.700766 | 3 | 0 | 3 |
| FRS2 | GYG2 | 0.694569 | 4 | 0 | 4 |
| FRS2 | SNX13 | 0.694455 | 3 | 0 | 3 |
| FRS2 | AFF3 | 0.692495 | 3 | 0 | 3 |
| FRS2 | WDFY3 | 0.691048 | 3 | 0 | 3 |
| FRS2 | SEC22B | 0.688594 | 3 | 0 | 3 |
| FRS2 | CNOT2 | 0.686653 | 4 | 0 | 3 |
For details and further investigation, click here