| Full name: glutamate decarboxylase 1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 2q31.1 | ||
| Entrez ID: 2571 | HGNC ID: HGNC:4092 | Ensembl Gene: ENSG00000128683 | OMIM ID: 605363 |
| Drug and gene relationship at DGIdb | |||
Expression of GAD1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GAD1 | 2571 | 206669_at | -0.0629 | 0.8823 | |
| GSE20347 | GAD1 | 2571 | 206669_at | 0.2997 | 0.0254 | |
| GSE23400 | GAD1 | 2571 | 206670_s_at | 0.0331 | 0.4822 | |
| GSE26886 | GAD1 | 2571 | 206669_at | 1.6834 | 0.0000 | |
| GSE29001 | GAD1 | 2571 | 205278_at | -0.4454 | 0.6230 | |
| GSE38129 | GAD1 | 2571 | 206669_at | 0.5419 | 0.0002 | |
| GSE45670 | GAD1 | 2571 | 206669_at | 0.1860 | 0.3774 | |
| GSE53622 | GAD1 | 2571 | 41219 | 0.5611 | 0.0167 | |
| GSE53624 | GAD1 | 2571 | 40433 | 1.0673 | 0.0000 | |
| GSE63941 | GAD1 | 2571 | 206670_s_at | 2.6277 | 0.0123 | |
| GSE77861 | GAD1 | 2571 | 206669_at | -0.0916 | 0.3105 | |
| SRP219564 | GAD1 | 2571 | RNAseq | 1.3049 | 0.1757 | |
| TCGA | GAD1 | 2571 | RNAseq | 2.1834 | 0.0000 |
Upregulated datasets: 4; Downregulated datasets: 0.
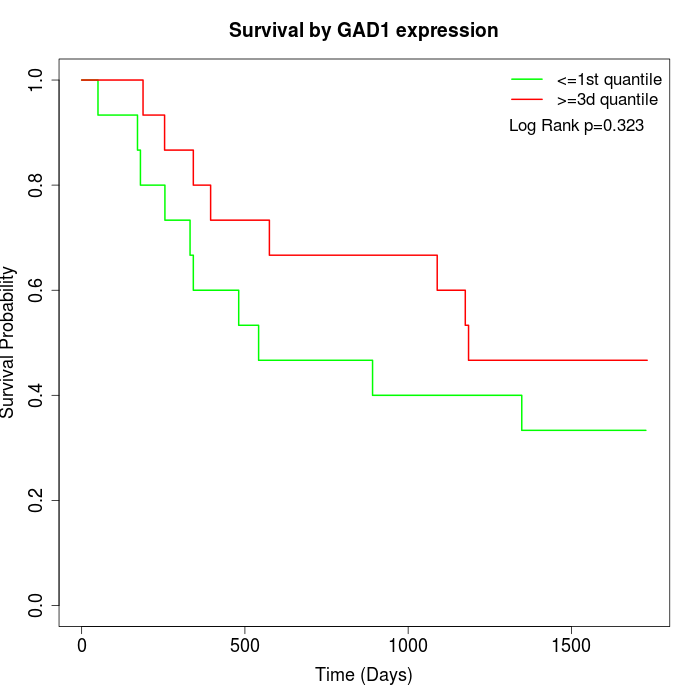
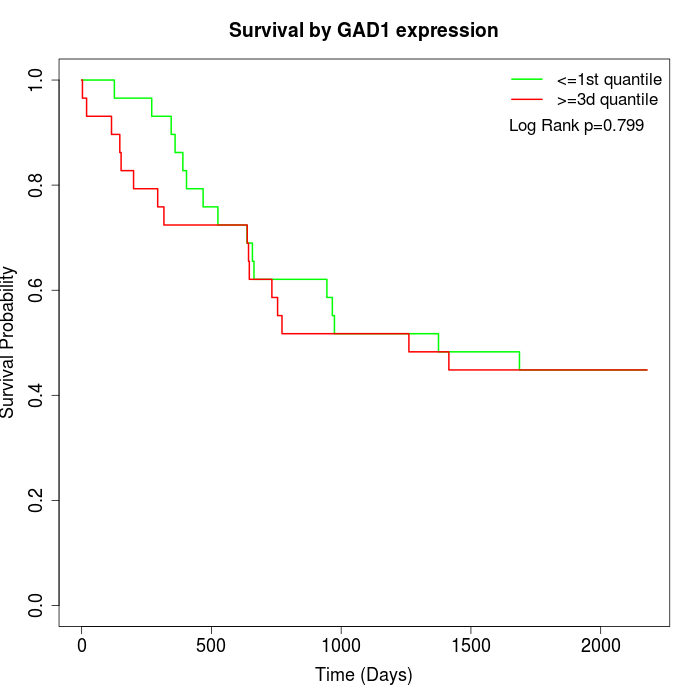
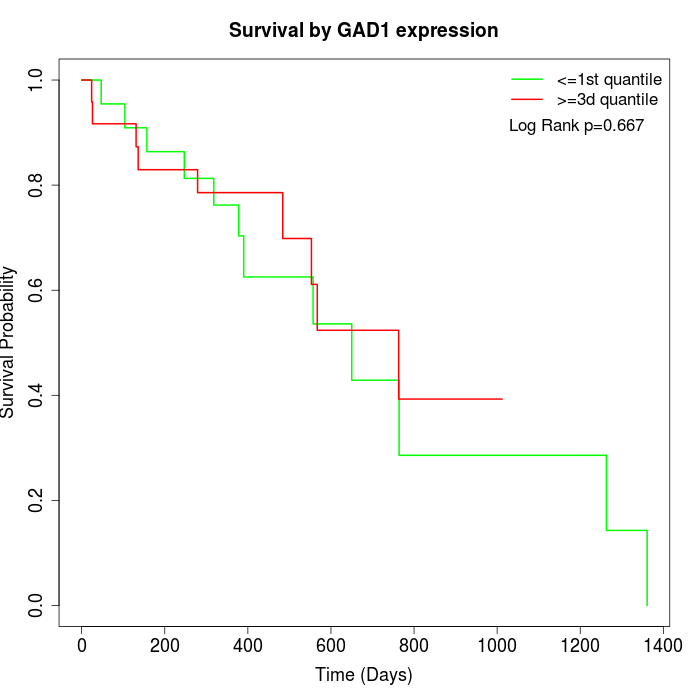
Survival by GAD1 expression:
Note: Click image to view full size file.
Copy number change of GAD1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GAD1 | 2571 | 8 | 0 | 22 | |
| GSE20123 | GAD1 | 2571 | 8 | 0 | 22 | |
| GSE43470 | GAD1 | 2571 | 5 | 1 | 37 | |
| GSE46452 | GAD1 | 2571 | 1 | 4 | 54 | |
| GSE47630 | GAD1 | 2571 | 5 | 3 | 32 | |
| GSE54993 | GAD1 | 2571 | 0 | 5 | 65 | |
| GSE54994 | GAD1 | 2571 | 11 | 3 | 39 | |
| GSE60625 | GAD1 | 2571 | 0 | 3 | 8 | |
| GSE74703 | GAD1 | 2571 | 4 | 1 | 31 | |
| GSE74704 | GAD1 | 2571 | 5 | 0 | 15 | |
| TCGA | GAD1 | 2571 | 27 | 7 | 62 |
Total number of gains: 74; Total number of losses: 27; Total Number of normals: 387.
Somatic mutations of GAD1:
Generating mutation plots.
Highly correlated genes for GAD1:
Showing top 20/47 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GAD1 | NOTUM | 0.666466 | 3 | 0 | 3 |
| GAD1 | FLT4 | 0.651603 | 3 | 0 | 3 |
| GAD1 | AMER3 | 0.650545 | 3 | 0 | 3 |
| GAD1 | RPS17 | 0.629805 | 3 | 0 | 3 |
| GAD1 | EGLN2 | 0.616198 | 3 | 0 | 3 |
| GAD1 | CARD10 | 0.615763 | 3 | 0 | 3 |
| GAD1 | COL26A1 | 0.615676 | 3 | 0 | 3 |
| GAD1 | SP5 | 0.613728 | 4 | 0 | 3 |
| GAD1 | CNTD1 | 0.594291 | 3 | 0 | 3 |
| GAD1 | GALP | 0.589596 | 4 | 0 | 3 |
| GAD1 | DPPA2 | 0.583697 | 3 | 0 | 3 |
| GAD1 | ATP8A2 | 0.578496 | 3 | 0 | 3 |
| GAD1 | TNNI3 | 0.573882 | 4 | 0 | 3 |
| GAD1 | MYT1 | 0.570023 | 3 | 0 | 3 |
| GAD1 | ELAVL3 | 0.567843 | 4 | 0 | 3 |
| GAD1 | GLTPD2 | 0.565816 | 4 | 0 | 3 |
| GAD1 | ADAM8 | 0.563574 | 3 | 0 | 3 |
| GAD1 | ADAM11 | 0.561583 | 4 | 0 | 3 |
| GAD1 | SYCE3 | 0.559508 | 4 | 0 | 3 |
| GAD1 | PRX | 0.558956 | 3 | 0 | 3 |
For details and further investigation, click here