| Full name: Sp5 transcription factor | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 2q31.1 | ||
| Entrez ID: 389058 | HGNC ID: HGNC:14529 | Ensembl Gene: ENSG00000204335 | OMIM ID: 609391 |
| Drug and gene relationship at DGIdb | |||
Expression of SP5:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SP5 | 389058 | 235845_at | -0.2534 | 0.2706 | |
| GSE26886 | SP5 | 389058 | 235845_at | 0.4616 | 0.0211 | |
| GSE45670 | SP5 | 389058 | 235845_at | -0.4320 | 0.0032 | |
| GSE53622 | SP5 | 389058 | 106003 | -1.5101 | 0.0000 | |
| GSE53624 | SP5 | 389058 | 106003 | -1.9757 | 0.0000 | |
| GSE63941 | SP5 | 389058 | 235845_at | 0.8649 | 0.0079 | |
| GSE77861 | SP5 | 389058 | 235845_at | -0.0099 | 0.9350 | |
| GSE97050 | SP5 | 389058 | A_23_P135381 | -0.0963 | 0.6793 | |
| SRP133303 | SP5 | 389058 | RNAseq | -3.3042 | 0.0000 | |
| TCGA | SP5 | 389058 | RNAseq | -1.9270 | 0.0002 |
Upregulated datasets: 0; Downregulated datasets: 4.
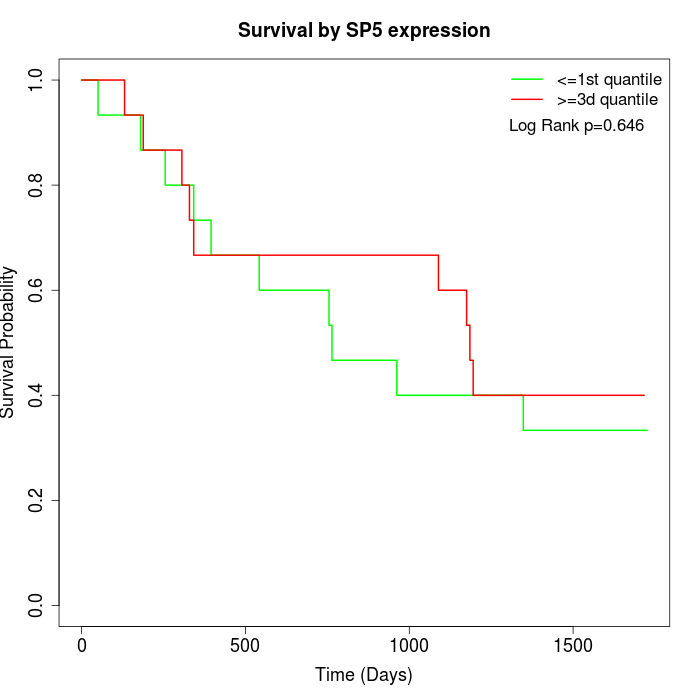
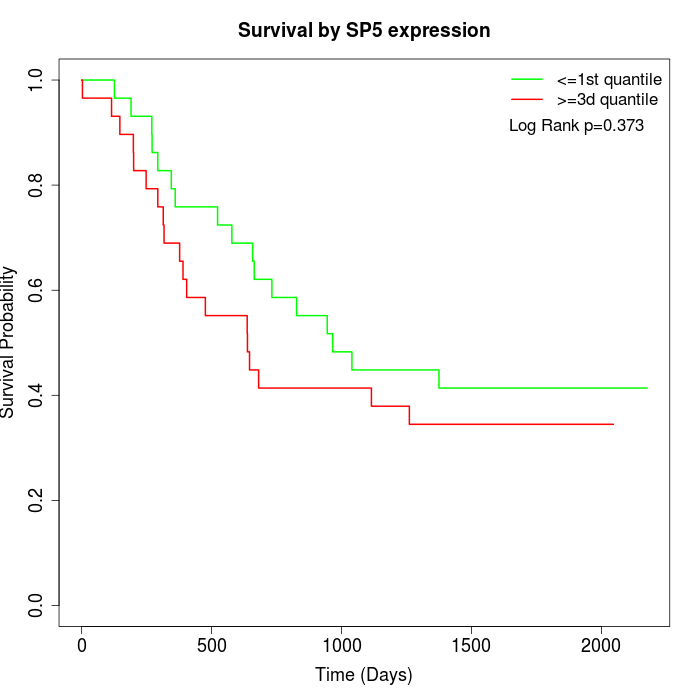
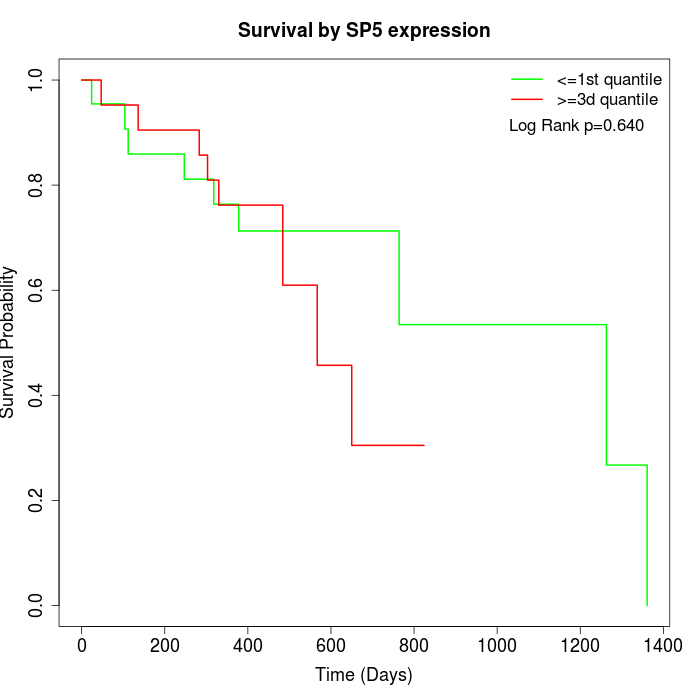
Survival by SP5 expression:
Note: Click image to view full size file.
Copy number change of SP5:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SP5 | 389058 | 8 | 0 | 22 | |
| GSE20123 | SP5 | 389058 | 8 | 0 | 22 | |
| GSE43470 | SP5 | 389058 | 5 | 1 | 37 | |
| GSE46452 | SP5 | 389058 | 1 | 4 | 54 | |
| GSE47630 | SP5 | 389058 | 5 | 3 | 32 | |
| GSE54993 | SP5 | 389058 | 0 | 5 | 65 | |
| GSE54994 | SP5 | 389058 | 11 | 3 | 39 | |
| GSE60625 | SP5 | 389058 | 0 | 3 | 8 | |
| GSE74703 | SP5 | 389058 | 4 | 1 | 31 | |
| GSE74704 | SP5 | 389058 | 5 | 0 | 15 | |
| TCGA | SP5 | 389058 | 25 | 9 | 62 |
Total number of gains: 72; Total number of losses: 29; Total Number of normals: 387.
Somatic mutations of SP5:
Generating mutation plots.
Highly correlated genes for SP5:
Showing top 20/47 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SP5 | FGF3 | 0.670391 | 3 | 0 | 3 |
| SP5 | GAD1 | 0.613728 | 4 | 0 | 3 |
| SP5 | FGF20 | 0.612131 | 4 | 0 | 3 |
| SP5 | SOX7 | 0.608249 | 3 | 0 | 3 |
| SP5 | SLITRK4 | 0.604522 | 3 | 0 | 3 |
| SP5 | STAB2 | 0.596605 | 4 | 0 | 3 |
| SP5 | ZNF503-AS1 | 0.58534 | 5 | 0 | 4 |
| SP5 | LIFR | 0.585137 | 4 | 0 | 3 |
| SP5 | NFIA | 0.572006 | 3 | 0 | 3 |
| SP5 | LGI1 | 0.569797 | 4 | 0 | 3 |
| SP5 | TFAP2B | 0.568615 | 4 | 0 | 3 |
| SP5 | SLC18A2 | 0.567077 | 5 | 0 | 4 |
| SP5 | SLC5A2 | 0.566867 | 3 | 0 | 3 |
| SP5 | OR10C1 | 0.565062 | 3 | 0 | 3 |
| SP5 | NCOA1 | 0.563651 | 4 | 0 | 4 |
| SP5 | ARPC2 | 0.562224 | 4 | 0 | 3 |
| SP5 | PROM1 | 0.557996 | 5 | 0 | 4 |
| SP5 | PDK4 | 0.55793 | 4 | 0 | 4 |
| SP5 | UTRN | 0.557182 | 4 | 0 | 4 |
| SP5 | FCER1A | 0.553717 | 5 | 0 | 4 |
For details and further investigation, click here