| Full name: germinal center associated signaling and motility like | Alias Symbol: FLJ44728 | ||
| Type: protein-coding gene | Cytoband: 1q44 | ||
| Entrez ID: 148823 | HGNC ID: HGNC:29583 | Ensembl Gene: ENSG00000169224 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of GCSAML:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GCSAML | 148823 | 1552908_at | -0.0240 | 0.9467 | |
| GSE26886 | GCSAML | 148823 | 1552908_at | 0.0567 | 0.5830 | |
| GSE45670 | GCSAML | 148823 | 1552908_at | -0.1299 | 0.2928 | |
| GSE53622 | GCSAML | 148823 | 6793 | -1.7761 | 0.0000 | |
| GSE53624 | GCSAML | 148823 | 6793 | -1.1627 | 0.0000 | |
| GSE63941 | GCSAML | 148823 | 1552908_at | -0.0183 | 0.8827 | |
| GSE77861 | GCSAML | 148823 | 1552908_at | -0.1046 | 0.2587 | |
| SRP064894 | GCSAML | 148823 | RNAseq | -0.6878 | 0.0081 | |
| SRP133303 | GCSAML | 148823 | RNAseq | 0.0424 | 0.7577 | |
| SRP159526 | GCSAML | 148823 | RNAseq | 0.3687 | 0.3131 | |
| SRP193095 | GCSAML | 148823 | RNAseq | -0.0030 | 0.9793 | |
| SRP219564 | GCSAML | 148823 | RNAseq | -0.2556 | 0.6627 |
Upregulated datasets: 0; Downregulated datasets: 2.
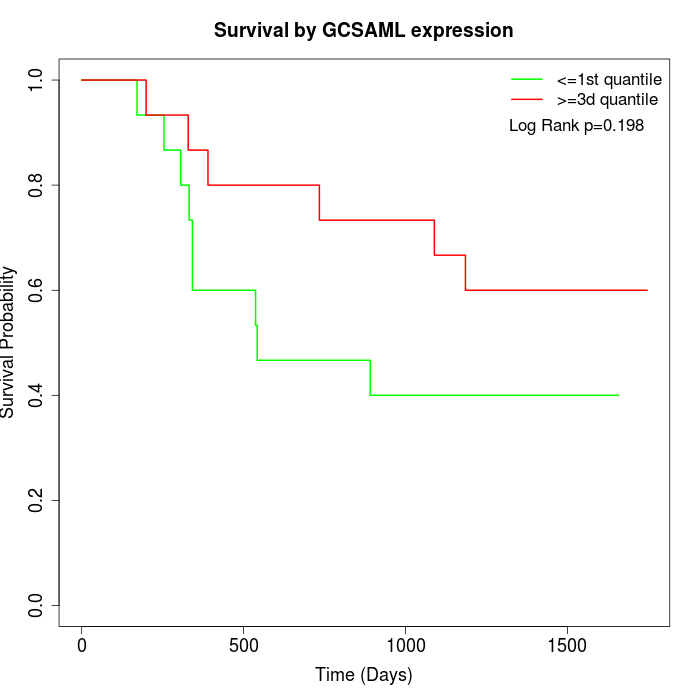
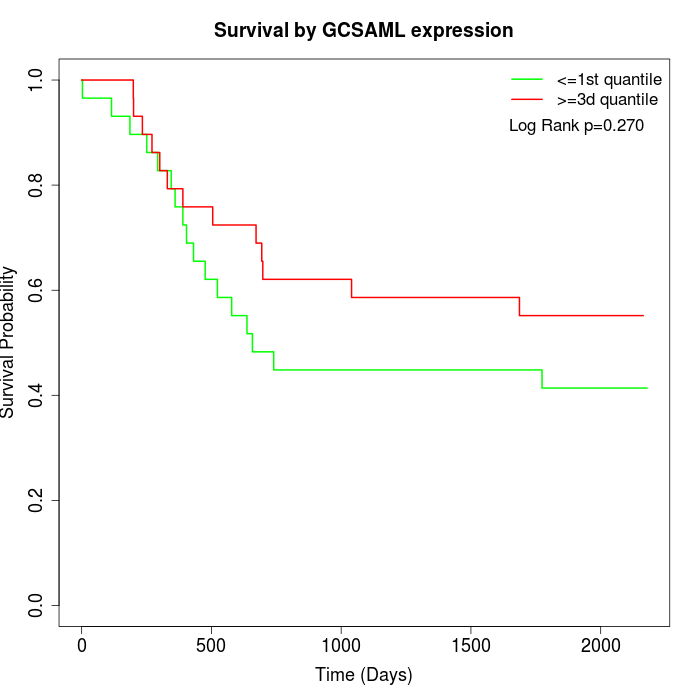
Survival by GCSAML expression:
Note: Click image to view full size file.
Copy number change of GCSAML:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GCSAML | 148823 | 7 | 1 | 22 | |
| GSE20123 | GCSAML | 148823 | 7 | 1 | 22 | |
| GSE43470 | GCSAML | 148823 | 4 | 3 | 36 | |
| GSE46452 | GCSAML | 148823 | 4 | 2 | 53 | |
| GSE47630 | GCSAML | 148823 | 15 | 1 | 24 | |
| GSE54993 | GCSAML | 148823 | 0 | 6 | 64 | |
| GSE54994 | GCSAML | 148823 | 18 | 0 | 35 | |
| GSE60625 | GCSAML | 148823 | 0 | 0 | 11 | |
| GSE74703 | GCSAML | 148823 | 4 | 1 | 31 | |
| GSE74704 | GCSAML | 148823 | 2 | 0 | 18 | |
| TCGA | GCSAML | 148823 | 45 | 4 | 47 |
Total number of gains: 106; Total number of losses: 19; Total Number of normals: 363.
Somatic mutations of GCSAML:
Generating mutation plots.
Highly correlated genes for GCSAML:
Showing top 20/102 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GCSAML | RECK | 0.68395 | 3 | 0 | 3 |
| GCSAML | LYVE1 | 0.682864 | 3 | 0 | 3 |
| GCSAML | F10 | 0.670512 | 3 | 0 | 3 |
| GCSAML | BMX | 0.669378 | 3 | 0 | 3 |
| GCSAML | GIMAP1 | 0.662092 | 3 | 0 | 3 |
| GCSAML | HPGDS | 0.658102 | 4 | 0 | 3 |
| GCSAML | PRSS35 | 0.655749 | 3 | 0 | 3 |
| GCSAML | C1QTNF2 | 0.653417 | 3 | 0 | 3 |
| GCSAML | CYYR1 | 0.652592 | 3 | 0 | 3 |
| GCSAML | MYRIP | 0.64666 | 4 | 0 | 4 |
| GCSAML | CMA1 | 0.64277 | 4 | 0 | 3 |
| GCSAML | SLC16A4 | 0.635268 | 3 | 0 | 3 |
| GCSAML | MMRN2 | 0.634527 | 3 | 0 | 3 |
| GCSAML | CYP2U1 | 0.633978 | 3 | 0 | 3 |
| GCSAML | C1QTNF7 | 0.63048 | 3 | 0 | 3 |
| GCSAML | GPD1L | 0.627869 | 3 | 0 | 3 |
| GCSAML | BHLHE22 | 0.627537 | 3 | 0 | 3 |
| GCSAML | SH3BGRL | 0.625659 | 3 | 0 | 3 |
| GCSAML | SHOC2 | 0.625513 | 3 | 0 | 3 |
| GCSAML | CPA3 | 0.621985 | 4 | 0 | 3 |
For details and further investigation, click here