| Full name: galactosidase beta 1 | Alias Symbol: EBP | ||
| Type: protein-coding gene | Cytoband: 3p22.3 | ||
| Entrez ID: 2720 | HGNC ID: HGNC:4298 | Ensembl Gene: ENSG00000170266 | OMIM ID: 611458 |
| Drug and gene relationship at DGIdb | |||
Expression of GLB1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | GLB1 | 2720 | 140724 | 0.2543 | 0.0036 | |
| GSE53624 | GLB1 | 2720 | 140724 | 0.3230 | 0.0003 | |
| GSE97050 | GLB1 | 2720 | A_23_P61531 | 0.2599 | 0.4386 | |
| SRP007169 | GLB1 | 2720 | RNAseq | 0.3471 | 0.3012 | |
| SRP008496 | GLB1 | 2720 | RNAseq | 0.5732 | 0.0437 | |
| SRP064894 | GLB1 | 2720 | RNAseq | 0.8395 | 0.0000 | |
| SRP133303 | GLB1 | 2720 | RNAseq | 0.3405 | 0.0143 | |
| SRP159526 | GLB1 | 2720 | RNAseq | 0.1534 | 0.6409 | |
| SRP193095 | GLB1 | 2720 | RNAseq | 0.1403 | 0.5416 | |
| SRP219564 | GLB1 | 2720 | RNAseq | 0.4980 | 0.1500 | |
| TCGA | GLB1 | 2720 | RNAseq | -0.1664 | 0.0005 |
Upregulated datasets: 0; Downregulated datasets: 0.
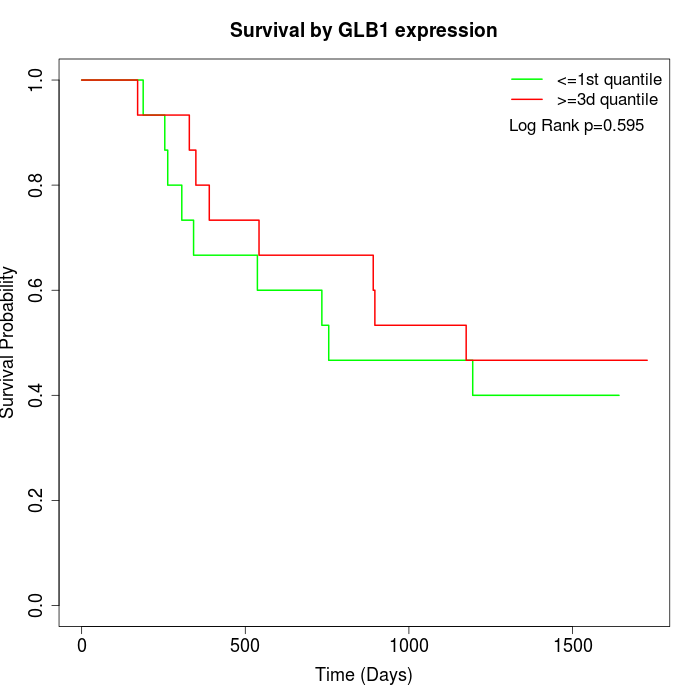
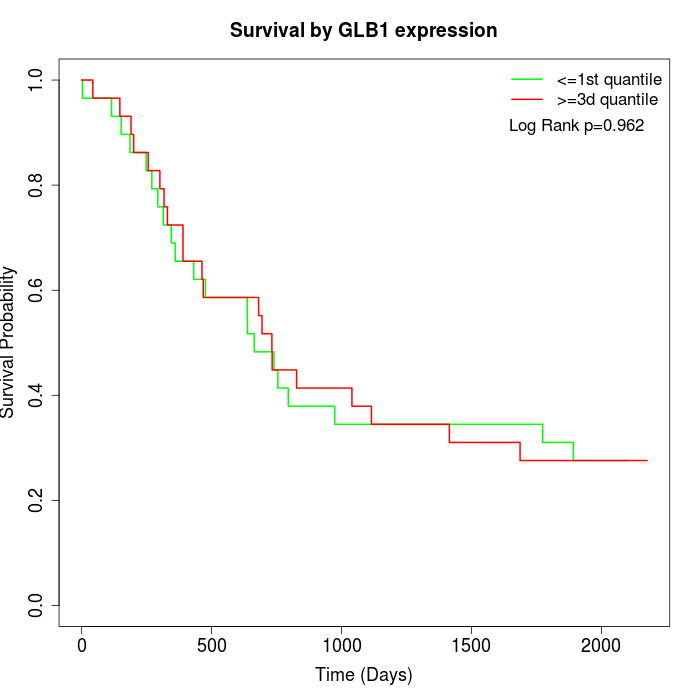
Survival by GLB1 expression:
Note: Click image to view full size file.
Copy number change of GLB1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GLB1 | 2720 | 0 | 18 | 12 | |
| GSE20123 | GLB1 | 2720 | 0 | 18 | 12 | |
| GSE43470 | GLB1 | 2720 | 1 | 19 | 23 | |
| GSE46452 | GLB1 | 2720 | 2 | 17 | 40 | |
| GSE47630 | GLB1 | 2720 | 1 | 25 | 14 | |
| GSE54993 | GLB1 | 2720 | 6 | 3 | 61 | |
| GSE54994 | GLB1 | 2720 | 1 | 32 | 20 | |
| GSE60625 | GLB1 | 2720 | 5 | 0 | 6 | |
| GSE74703 | GLB1 | 2720 | 1 | 16 | 19 | |
| GSE74704 | GLB1 | 2720 | 0 | 12 | 8 | |
| TCGA | GLB1 | 2720 | 1 | 69 | 26 |
Total number of gains: 18; Total number of losses: 229; Total Number of normals: 241.
Somatic mutations of GLB1:
Generating mutation plots.
Highly correlated genes for GLB1:
Showing top 20/37 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GLB1 | PLCL2 | 0.684727 | 3 | 0 | 3 |
| GLB1 | C2 | 0.676297 | 3 | 0 | 3 |
| GLB1 | IL10RA | 0.676139 | 3 | 0 | 3 |
| GLB1 | CYBB | 0.676029 | 3 | 0 | 3 |
| GLB1 | SLAMF8 | 0.67469 | 3 | 0 | 3 |
| GLB1 | LAIR2 | 0.667848 | 3 | 0 | 3 |
| GLB1 | PRDX4 | 0.665825 | 3 | 0 | 3 |
| GLB1 | CXorf21 | 0.65463 | 3 | 0 | 3 |
| GLB1 | CD300LF | 0.653819 | 3 | 0 | 3 |
| GLB1 | SDSL | 0.645757 | 3 | 0 | 3 |
| GLB1 | TBXAS1 | 0.642477 | 4 | 0 | 3 |
| GLB1 | TMEM150B | 0.639844 | 3 | 0 | 3 |
| GLB1 | IGFLR1 | 0.623268 | 3 | 0 | 3 |
| GLB1 | SLC7A7 | 0.615321 | 4 | 0 | 3 |
| GLB1 | AIF1 | 0.61353 | 3 | 0 | 3 |
| GLB1 | KLRD1 | 0.604679 | 3 | 0 | 3 |
| GLB1 | GPN2 | 0.601629 | 3 | 0 | 3 |
| GLB1 | CMTM7 | 0.595357 | 3 | 0 | 3 |
| GLB1 | FMNL1 | 0.594864 | 3 | 0 | 3 |
| GLB1 | TLR8 | 0.59427 | 3 | 0 | 3 |
For details and further investigation, click here