| Full name: complement C2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 6p21.33 | ||
| Entrez ID: 717 | HGNC ID: HGNC:1248 | Ensembl Gene: ENSG00000166278 | OMIM ID: 613927 |
| Drug and gene relationship at DGIdb | |||
C2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04610 | Complement and coagulation cascades | |
| hsa05133 | Pertussis | |
| hsa05150 | Staphylococcus aureus infection |
Expression of C2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | C2 | 717 | 203052_at | 0.2275 | 0.7542 | |
| GSE20347 | C2 | 717 | 203052_at | 0.1800 | 0.1310 | |
| GSE23400 | C2 | 717 | 203052_at | -0.0043 | 0.9416 | |
| GSE26886 | C2 | 717 | 203052_at | 0.1826 | 0.3581 | |
| GSE29001 | C2 | 717 | 203052_at | 0.1523 | 0.4396 | |
| GSE38129 | C2 | 717 | 203052_at | 0.1739 | 0.2445 | |
| GSE45670 | C2 | 717 | 203052_at | -0.1278 | 0.4625 | |
| GSE53622 | C2 | 717 | 92671 | 1.0996 | 0.0000 | |
| GSE53624 | C2 | 717 | 92671 | 1.2553 | 0.0000 | |
| GSE63941 | C2 | 717 | 203052_at | -0.2657 | 0.6219 | |
| GSE77861 | C2 | 717 | 203052_at | 0.0423 | 0.7876 | |
| GSE97050 | C2 | 717 | A_33_P3404601 | 0.5407 | 0.2312 | |
| SRP007169 | C2 | 717 | RNAseq | 1.7835 | 0.0166 | |
| SRP008496 | C2 | 717 | RNAseq | 2.3127 | 0.0000 | |
| SRP064894 | C2 | 717 | RNAseq | 1.0707 | 0.0042 | |
| SRP133303 | C2 | 717 | RNAseq | 0.9121 | 0.0003 | |
| SRP159526 | C2 | 717 | RNAseq | 0.9642 | 0.1255 | |
| SRP193095 | C2 | 717 | RNAseq | 1.2165 | 0.0000 | |
| SRP219564 | C2 | 717 | RNAseq | 2.6184 | 0.0003 | |
| TCGA | C2 | 717 | RNAseq | 0.2546 | 0.0460 |
Upregulated datasets: 7; Downregulated datasets: 0.
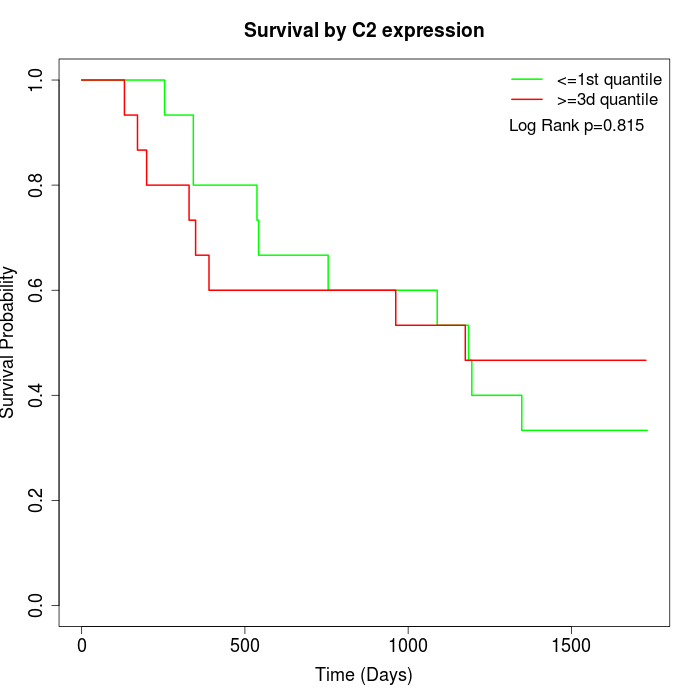
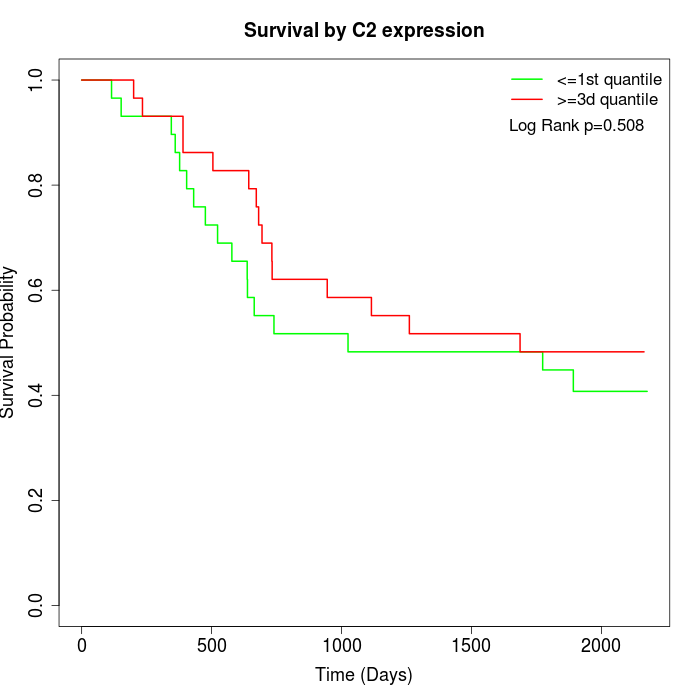
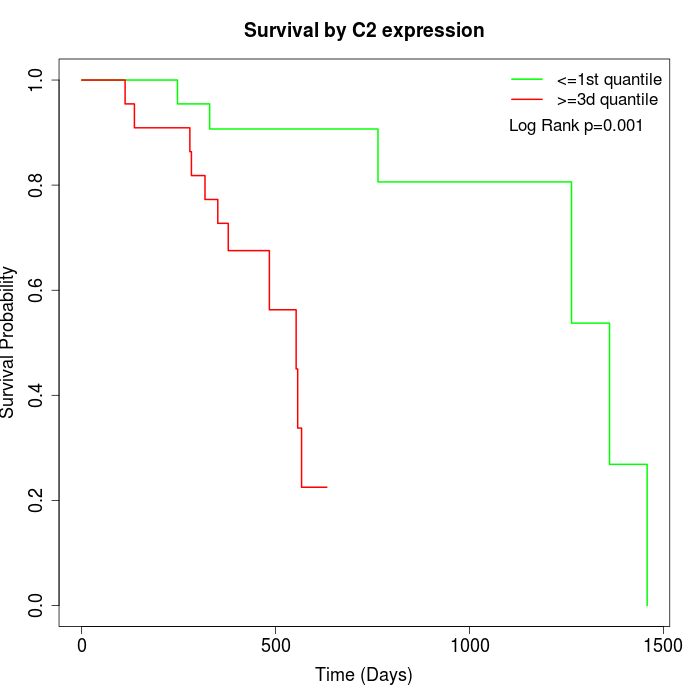
Survival by C2 expression:
Note: Click image to view full size file.
Copy number change of C2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | C2 | 717 | 5 | 1 | 24 | |
| GSE20123 | C2 | 717 | 5 | 1 | 24 | |
| GSE43470 | C2 | 717 | 5 | 1 | 37 | |
| GSE46452 | C2 | 717 | 1 | 10 | 48 | |
| GSE47630 | C2 | 717 | 7 | 4 | 29 | |
| GSE54993 | C2 | 717 | 2 | 1 | 67 | |
| GSE54994 | C2 | 717 | 11 | 4 | 38 | |
| GSE60625 | C2 | 717 | 0 | 1 | 10 | |
| GSE74703 | C2 | 717 | 5 | 0 | 31 | |
| GSE74704 | C2 | 717 | 2 | 0 | 18 | |
| TCGA | C2 | 717 | 16 | 16 | 64 |
Total number of gains: 59; Total number of losses: 39; Total Number of normals: 390.
Somatic mutations of C2:
Generating mutation plots.
Highly correlated genes for C2:
Showing top 20/468 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| C2 | SIGLEC12 | 0.707516 | 3 | 0 | 3 |
| C2 | PLXNC1 | 0.700657 | 7 | 0 | 7 |
| C2 | CYBB | 0.698892 | 9 | 0 | 9 |
| C2 | SDSL | 0.68909 | 4 | 0 | 4 |
| C2 | C1QA | 0.687574 | 10 | 0 | 10 |
| C2 | LCP2 | 0.686143 | 8 | 0 | 7 |
| C2 | ADAMDEC1 | 0.681549 | 7 | 0 | 6 |
| C2 | GLB1 | 0.676297 | 3 | 0 | 3 |
| C2 | LAIR1 | 0.67469 | 8 | 0 | 7 |
| C2 | CCL4L2 | 0.667427 | 3 | 0 | 3 |
| C2 | CDH11 | 0.664199 | 3 | 0 | 3 |
| C2 | SLAMF8 | 0.662497 | 12 | 0 | 10 |
| C2 | CMTM7 | 0.659511 | 6 | 0 | 5 |
| C2 | TRPV2 | 0.657625 | 11 | 0 | 9 |
| C2 | IDO1 | 0.65409 | 8 | 0 | 7 |
| C2 | TMEM119 | 0.652197 | 6 | 0 | 4 |
| C2 | MYLK-AS1 | 0.649627 | 3 | 0 | 3 |
| C2 | BCAT1 | 0.647313 | 4 | 0 | 4 |
| C2 | FGF18 | 0.647306 | 3 | 0 | 3 |
| C2 | TMEM200A | 0.646297 | 4 | 0 | 4 |
For details and further investigation, click here