| Full name: killer cell lectin like receptor D1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 12p13 | ||
| Entrez ID: 3824 | HGNC ID: HGNC:6378 | Ensembl Gene: ENSG00000134539 | OMIM ID: 602894 |
| Drug and gene relationship at DGIdb | |||
KLRD1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04612 | Antigen processing and presentation | |
| hsa04650 | Natural killer cell mediated cytotoxicity |
Expression of KLRD1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KLRD1 | 3824 | 210606_x_at | 0.3406 | 0.4748 | |
| GSE20347 | KLRD1 | 3824 | 207795_s_at | -0.1490 | 0.0348 | |
| GSE23400 | KLRD1 | 3824 | 210606_x_at | 0.0506 | 0.2616 | |
| GSE26886 | KLRD1 | 3824 | 207795_s_at | -0.3075 | 0.0076 | |
| GSE29001 | KLRD1 | 3824 | 210606_x_at | -0.1295 | 0.4912 | |
| GSE38129 | KLRD1 | 3824 | 207795_s_at | -0.1858 | 0.0213 | |
| GSE45670 | KLRD1 | 3824 | 210606_x_at | -0.1171 | 0.5784 | |
| GSE53622 | KLRD1 | 3824 | 100814 | 0.3155 | 0.1497 | |
| GSE53624 | KLRD1 | 3824 | 100814 | 0.4247 | 0.0676 | |
| GSE63941 | KLRD1 | 3824 | 207795_s_at | 0.0368 | 0.9249 | |
| GSE77861 | KLRD1 | 3824 | 207795_s_at | -0.2551 | 0.0264 | |
| GSE97050 | KLRD1 | 3824 | A_33_P3406196 | 0.1372 | 0.8341 | |
| SRP007169 | KLRD1 | 3824 | RNAseq | 1.0369 | 0.0260 | |
| SRP008496 | KLRD1 | 3824 | RNAseq | 1.9720 | 0.0000 | |
| SRP064894 | KLRD1 | 3824 | RNAseq | 0.0688 | 0.7387 | |
| SRP133303 | KLRD1 | 3824 | RNAseq | 0.2400 | 0.0260 | |
| SRP159526 | KLRD1 | 3824 | RNAseq | 0.5416 | 0.1200 | |
| SRP193095 | KLRD1 | 3824 | RNAseq | 0.2495 | 0.0439 | |
| SRP219564 | KLRD1 | 3824 | RNAseq | 0.5028 | 0.1280 | |
| TCGA | KLRD1 | 3824 | RNAseq | 0.1255 | 0.6621 |
Upregulated datasets: 2; Downregulated datasets: 0.
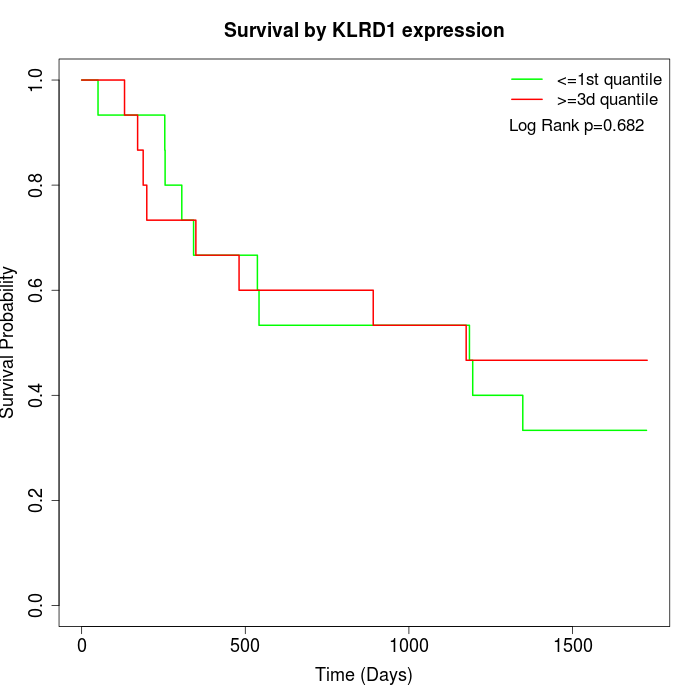
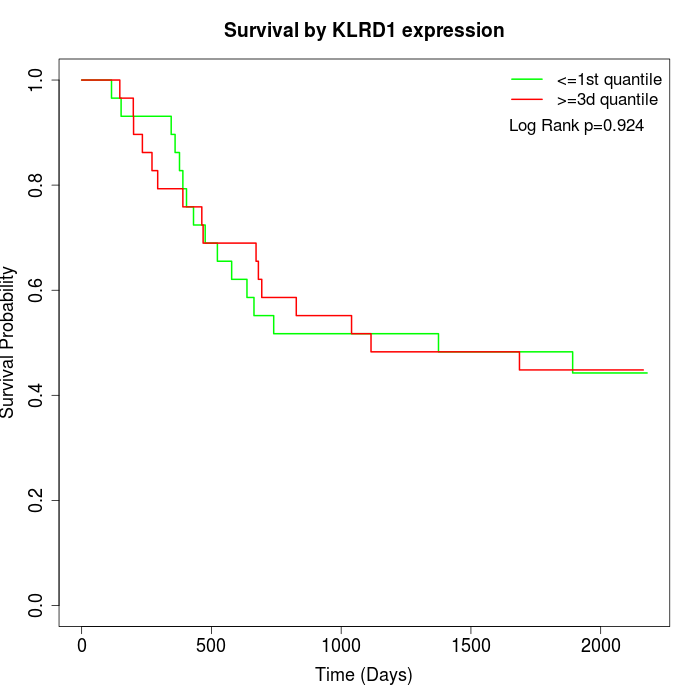
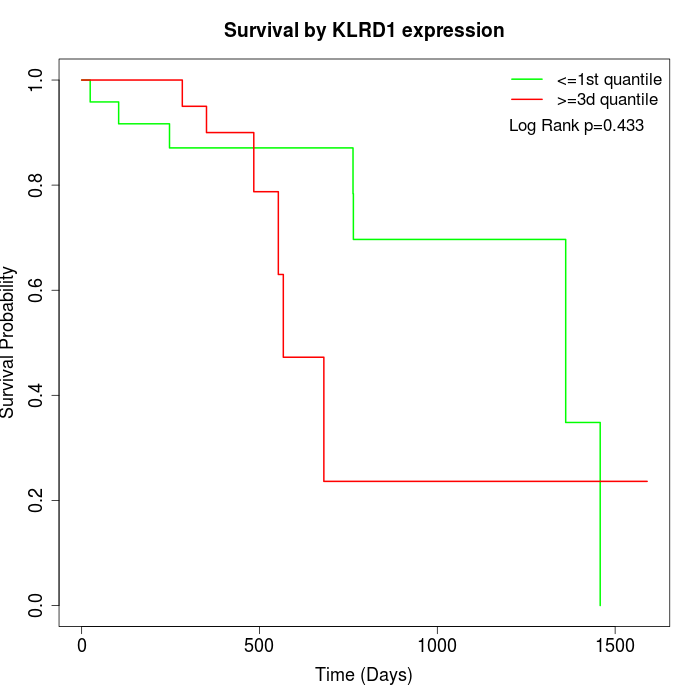
Survival by KLRD1 expression:
Note: Click image to view full size file.
Copy number change of KLRD1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KLRD1 | 3824 | 6 | 2 | 22 | |
| GSE20123 | KLRD1 | 3824 | 6 | 2 | 22 | |
| GSE43470 | KLRD1 | 3824 | 9 | 3 | 31 | |
| GSE46452 | KLRD1 | 3824 | 10 | 1 | 48 | |
| GSE47630 | KLRD1 | 3824 | 14 | 2 | 24 | |
| GSE54993 | KLRD1 | 3824 | 1 | 10 | 59 | |
| GSE54994 | KLRD1 | 3824 | 9 | 2 | 42 | |
| GSE60625 | KLRD1 | 3824 | 0 | 1 | 10 | |
| GSE74703 | KLRD1 | 3824 | 9 | 2 | 25 | |
| GSE74704 | KLRD1 | 3824 | 4 | 1 | 15 | |
| TCGA | KLRD1 | 3824 | 39 | 6 | 51 |
Total number of gains: 107; Total number of losses: 32; Total Number of normals: 349.
Somatic mutations of KLRD1:
Generating mutation plots.
Highly correlated genes for KLRD1:
Showing top 20/263 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KLRD1 | TMEM150B | 0.689223 | 4 | 0 | 4 |
| KLRD1 | MANF | 0.677763 | 3 | 0 | 3 |
| KLRD1 | GMFG | 0.675368 | 5 | 0 | 4 |
| KLRD1 | SIGLEC10 | 0.674654 | 6 | 0 | 5 |
| KLRD1 | CYBB | 0.674587 | 7 | 0 | 6 |
| KLRD1 | CAMK4 | 0.663551 | 5 | 0 | 5 |
| KLRD1 | CD4 | 0.653319 | 4 | 0 | 4 |
| KLRD1 | GZMA | 0.652654 | 6 | 0 | 6 |
| KLRD1 | TIGIT | 0.648493 | 5 | 0 | 4 |
| KLRD1 | FMNL1 | 0.648208 | 4 | 0 | 4 |
| KLRD1 | NKG7 | 0.646899 | 8 | 0 | 6 |
| KLRD1 | CRTAM | 0.645523 | 4 | 0 | 3 |
| KLRD1 | GDF5 | 0.644115 | 4 | 0 | 3 |
| KLRD1 | CCL4L2 | 0.640279 | 3 | 0 | 3 |
| KLRD1 | IGSF6 | 0.637729 | 5 | 0 | 4 |
| KLRD1 | TRIM25 | 0.635145 | 3 | 0 | 3 |
| KLRD1 | SAMD3 | 0.634123 | 5 | 0 | 4 |
| KLRD1 | TRANK1 | 0.628657 | 5 | 0 | 4 |
| KLRD1 | SAMSN1 | 0.626646 | 6 | 0 | 5 |
| KLRD1 | CD27 | 0.626596 | 3 | 0 | 3 |
For details and further investigation, click here