| Full name: glycine decarboxylase | Alias Symbol: GCSP|NKH | ||
| Type: protein-coding gene | Cytoband: 9p24.1 | ||
| Entrez ID: 2731 | HGNC ID: HGNC:4313 | Ensembl Gene: ENSG00000178445 | OMIM ID: 238300 |
| Drug and gene relationship at DGIdb | |||
Expression of GLDC:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GLDC | 2731 | 204836_at | 1.6334 | 0.3212 | |
| GSE20347 | GLDC | 2731 | 204836_at | 0.4850 | 0.0853 | |
| GSE23400 | GLDC | 2731 | 204836_at | 0.4176 | 0.0014 | |
| GSE26886 | GLDC | 2731 | 204836_at | -0.5446 | 0.1869 | |
| GSE29001 | GLDC | 2731 | 204836_at | 0.7721 | 0.1602 | |
| GSE38129 | GLDC | 2731 | 204836_at | 0.4454 | 0.0525 | |
| GSE45670 | GLDC | 2731 | 204836_at | 1.8190 | 0.0001 | |
| GSE53622 | GLDC | 2731 | 49378 | 0.4963 | 0.0125 | |
| GSE53624 | GLDC | 2731 | 49378 | 0.6780 | 0.0002 | |
| GSE63941 | GLDC | 2731 | 204836_at | 0.2982 | 0.6753 | |
| GSE77861 | GLDC | 2731 | 204836_at | -0.0605 | 0.7589 | |
| GSE97050 | GLDC | 2731 | A_23_P123596 | 0.6186 | 0.2035 | |
| SRP133303 | GLDC | 2731 | RNAseq | -0.6591 | 0.0771 | |
| SRP159526 | GLDC | 2731 | RNAseq | 0.1483 | 0.7703 | |
| SRP219564 | GLDC | 2731 | RNAseq | 0.0639 | 0.9413 | |
| TCGA | GLDC | 2731 | RNAseq | 1.4081 | 0.0028 |
Upregulated datasets: 2; Downregulated datasets: 0.
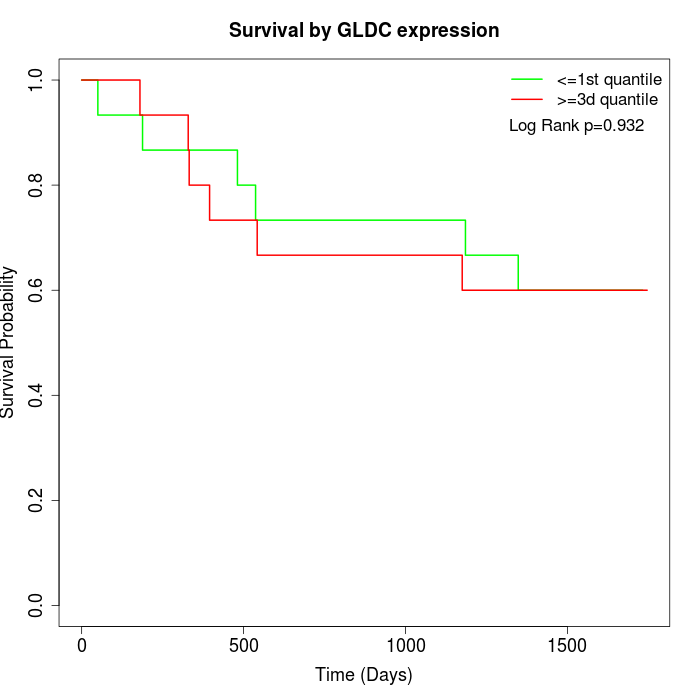
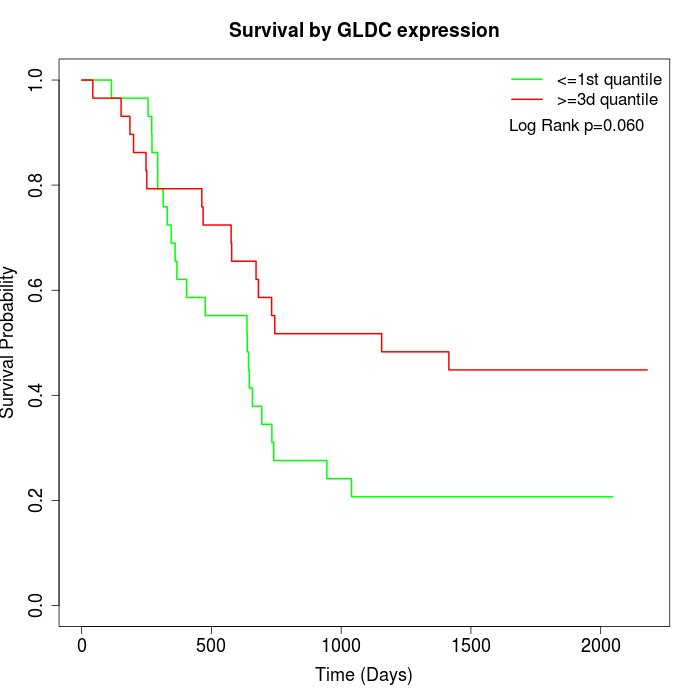
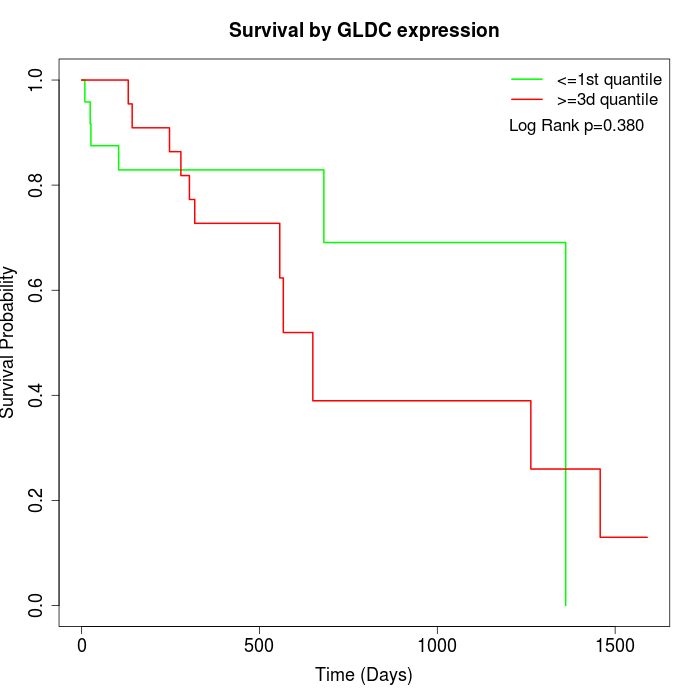
Survival by GLDC expression:
Note: Click image to view full size file.
Copy number change of GLDC:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GLDC | 2731 | 1 | 17 | 12 | |
| GSE20123 | GLDC | 2731 | 1 | 17 | 12 | |
| GSE43470 | GLDC | 2731 | 5 | 11 | 27 | |
| GSE46452 | GLDC | 2731 | 2 | 23 | 34 | |
| GSE47630 | GLDC | 2731 | 5 | 26 | 9 | |
| GSE54993 | GLDC | 2731 | 6 | 3 | 61 | |
| GSE54994 | GLDC | 2731 | 9 | 16 | 28 | |
| GSE60625 | GLDC | 2731 | 0 | 2 | 9 | |
| GSE74703 | GLDC | 2731 | 5 | 8 | 23 | |
| GSE74704 | GLDC | 2731 | 0 | 13 | 7 | |
| TCGA | GLDC | 2731 | 14 | 51 | 31 |
Total number of gains: 48; Total number of losses: 187; Total Number of normals: 253.
Somatic mutations of GLDC:
Generating mutation plots.
Highly correlated genes for GLDC:
Showing top 20/46 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GLDC | CD300LF | 0.797713 | 3 | 0 | 3 |
| GLDC | LRFN1 | 0.70692 | 3 | 0 | 3 |
| GLDC | ASB6 | 0.695712 | 3 | 0 | 3 |
| GLDC | DENND1C | 0.670608 | 4 | 0 | 3 |
| GLDC | POLR3G | 0.658982 | 7 | 0 | 6 |
| GLDC | MAK | 0.655033 | 3 | 0 | 3 |
| GLDC | KLHDC7B | 0.652774 | 3 | 0 | 3 |
| GLDC | PXT1 | 0.645331 | 3 | 0 | 3 |
| GLDC | EPHB2 | 0.644315 | 3 | 0 | 3 |
| GLDC | ZNF283 | 0.639062 | 3 | 0 | 3 |
| GLDC | LHFPL5 | 0.635871 | 4 | 0 | 3 |
| GLDC | ZNF335 | 0.630872 | 3 | 0 | 3 |
| GLDC | HTR6 | 0.628451 | 3 | 0 | 3 |
| GLDC | CLEC5A | 0.61859 | 3 | 0 | 3 |
| GLDC | MAGEB2 | 0.615349 | 3 | 0 | 3 |
| GLDC | TYMP | 0.593411 | 7 | 0 | 6 |
| GLDC | CHMP1A | 0.592437 | 4 | 0 | 3 |
| GLDC | MRPS17 | 0.590053 | 4 | 0 | 3 |
| GLDC | DUSP9 | 0.589358 | 6 | 0 | 3 |
| GLDC | TXNL4B | 0.586871 | 3 | 0 | 3 |
For details and further investigation, click here