| Full name: GLIS family zinc finger 1 | Alias Symbol: FLJ36155 | ||
| Type: protein-coding gene | Cytoband: 1p32.3 | ||
| Entrez ID: 148979 | HGNC ID: HGNC:29525 | Ensembl Gene: ENSG00000174332 | OMIM ID: 610378 |
| Drug and gene relationship at DGIdb | |||
Expression of GLIS1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GLIS1 | 148979 | 244128_x_at | -0.1743 | 0.6545 | |
| GSE26886 | GLIS1 | 148979 | 244128_x_at | 0.7884 | 0.0069 | |
| GSE45670 | GLIS1 | 148979 | 244128_x_at | 0.1185 | 0.6389 | |
| GSE53622 | GLIS1 | 148979 | 27889 | 0.2617 | 0.2600 | |
| GSE53624 | GLIS1 | 148979 | 27889 | 0.3291 | 0.0141 | |
| GSE63941 | GLIS1 | 148979 | 244128_x_at | -0.4639 | 0.1948 | |
| GSE77861 | GLIS1 | 148979 | 244128_x_at | 0.0282 | 0.8572 | |
| GSE97050 | GLIS1 | 148979 | A_23_P316612 | -0.1097 | 0.5811 | |
| SRP133303 | GLIS1 | 148979 | RNAseq | 0.2942 | 0.4726 | |
| SRP159526 | GLIS1 | 148979 | RNAseq | 0.3854 | 0.4622 | |
| SRP219564 | GLIS1 | 148979 | RNAseq | 2.0678 | 0.0107 | |
| TCGA | GLIS1 | 148979 | RNAseq | 0.3982 | 0.1545 |
Upregulated datasets: 1; Downregulated datasets: 0.
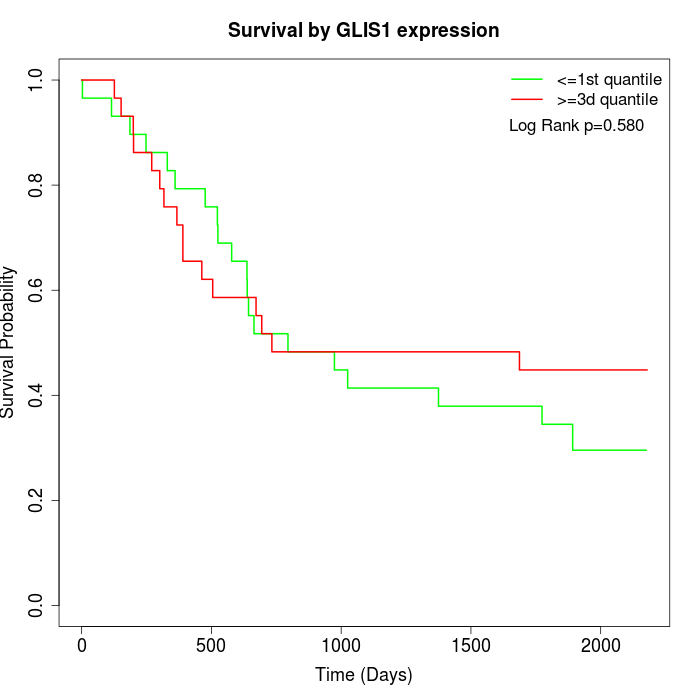
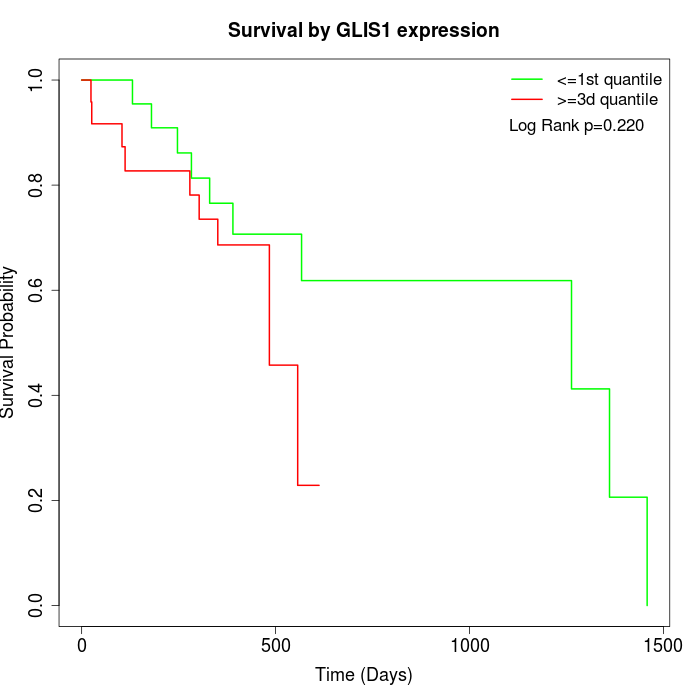
Survival by GLIS1 expression:
Note: Click image to view full size file.
Copy number change of GLIS1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GLIS1 | 148979 | 3 | 4 | 23 | |
| GSE20123 | GLIS1 | 148979 | 3 | 3 | 24 | |
| GSE43470 | GLIS1 | 148979 | 6 | 2 | 35 | |
| GSE46452 | GLIS1 | 148979 | 2 | 1 | 56 | |
| GSE47630 | GLIS1 | 148979 | 8 | 4 | 28 | |
| GSE54993 | GLIS1 | 148979 | 0 | 1 | 69 | |
| GSE54994 | GLIS1 | 148979 | 11 | 2 | 40 | |
| GSE60625 | GLIS1 | 148979 | 0 | 0 | 11 | |
| GSE74703 | GLIS1 | 148979 | 4 | 1 | 31 | |
| GSE74704 | GLIS1 | 148979 | 2 | 0 | 18 | |
| TCGA | GLIS1 | 148979 | 10 | 18 | 68 |
Total number of gains: 49; Total number of losses: 36; Total Number of normals: 403.
Somatic mutations of GLIS1:
Generating mutation plots.
Highly correlated genes for GLIS1:
Showing top 20/27 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GLIS1 | NPAS3 | 0.665884 | 3 | 0 | 3 |
| GLIS1 | KATNAL1 | 0.643534 | 3 | 0 | 3 |
| GLIS1 | CCNA1 | 0.625259 | 3 | 0 | 3 |
| GLIS1 | CHRNA4 | 0.611019 | 3 | 0 | 3 |
| GLIS1 | LIM2 | 0.587398 | 3 | 0 | 3 |
| GLIS1 | MAMLD1 | 0.568683 | 3 | 0 | 3 |
| GLIS1 | NEK10 | 0.556958 | 4 | 0 | 3 |
| GLIS1 | AIFM3 | 0.551923 | 3 | 0 | 3 |
| GLIS1 | GPR146 | 0.549138 | 4 | 0 | 3 |
| GLIS1 | SFXN3 | 0.549076 | 4 | 0 | 3 |
| GLIS1 | GPN2 | 0.545902 | 4 | 0 | 3 |
| GLIS1 | TSPAN10 | 0.543759 | 4 | 0 | 3 |
| GLIS1 | ARL4D | 0.543546 | 4 | 0 | 3 |
| GLIS1 | IGFL2 | 0.541782 | 7 | 0 | 3 |
| GLIS1 | LDB1 | 0.541555 | 4 | 0 | 3 |
| GLIS1 | RTBDN | 0.541344 | 5 | 0 | 4 |
| GLIS1 | ZNF319 | 0.538509 | 4 | 0 | 3 |
| GLIS1 | POU3F1 | 0.532416 | 6 | 0 | 3 |
| GLIS1 | PRKG1 | 0.528496 | 5 | 0 | 3 |
| GLIS1 | SNORD114-3 | 0.525131 | 4 | 0 | 3 |
For details and further investigation, click here